Pcr Primer Direction
They are synthesized chemically by joining nucleotides together.
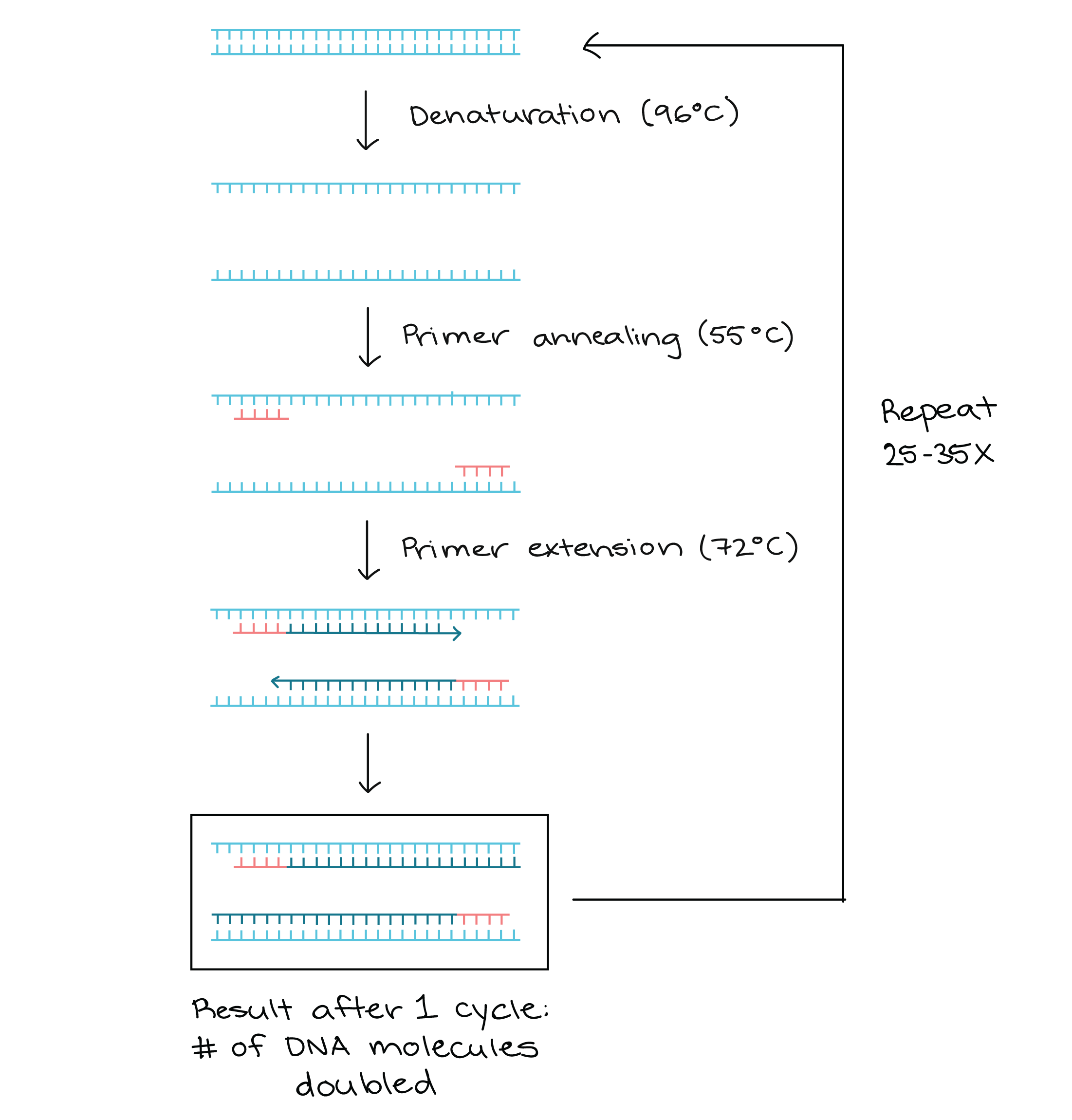
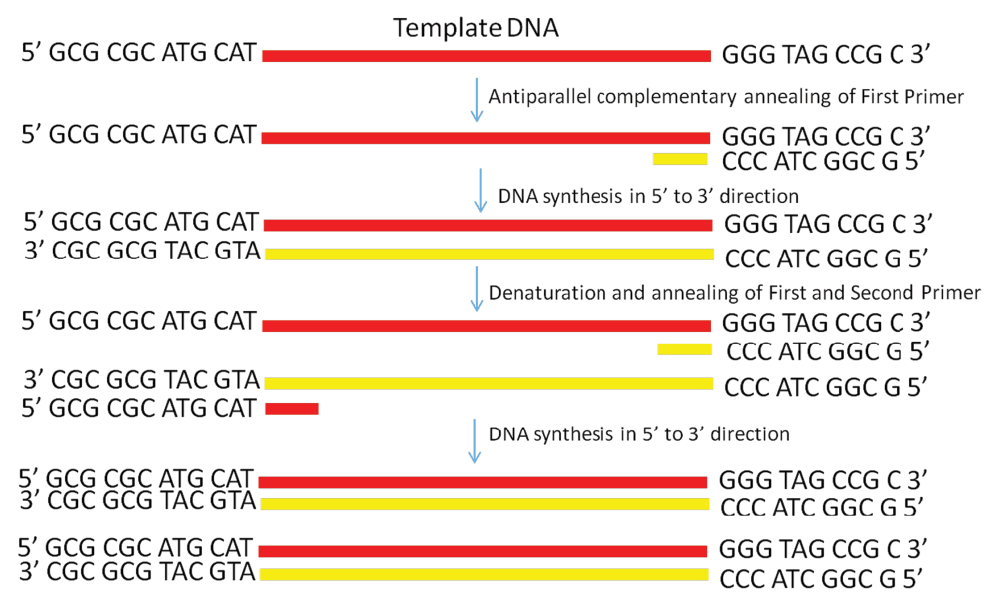
Pcr primer direction. Synthesis always occurs from the 5' to the 3' direction on each primer. And so when you cool it back down, the primer attaches, and then you heat it back up, not quite to the 96 degrees Celsius, but to the 72 degrees Celsius, where you extend those. After -25 cycles of PCR, one primer is exhausted.
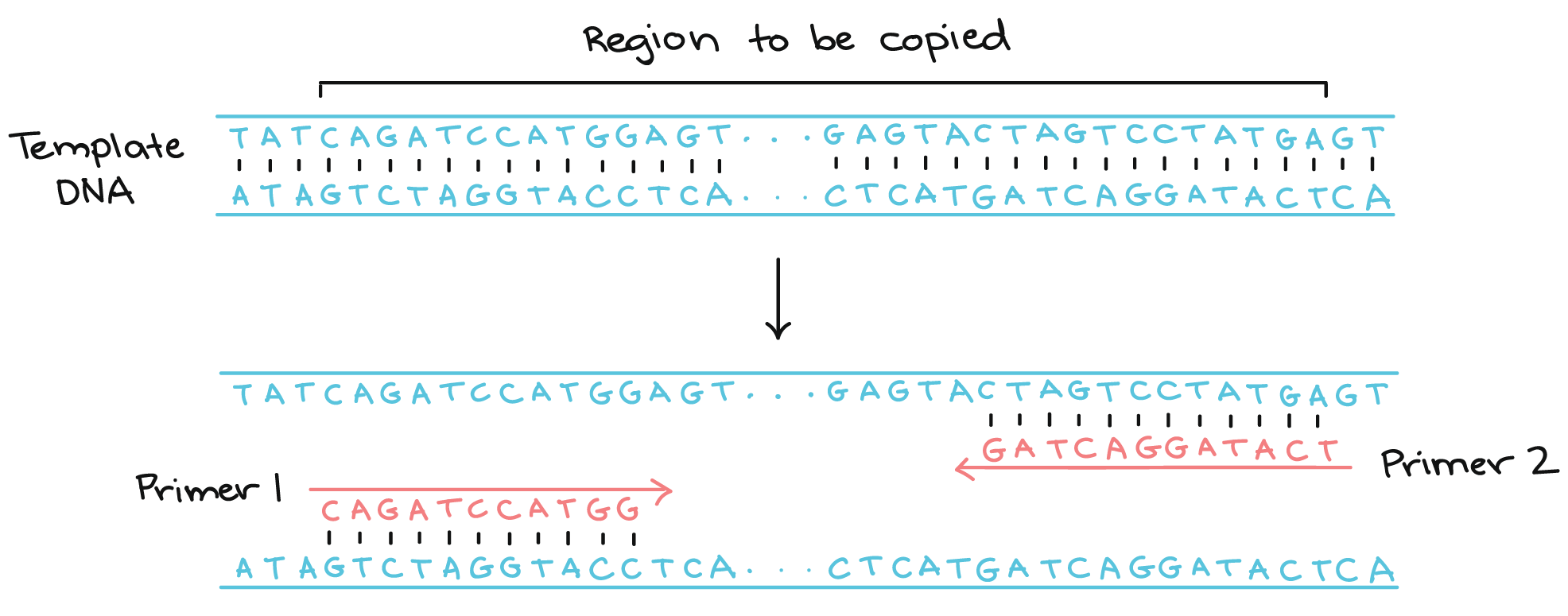
How to Make Primers for PCR The direction of both forward and reverse primer should be 5′ to 3′. The result is a brand new strand of DNA and a double-stranded molecule of DNA. In a PCR reaction, the experimenter determines the region of DNA that will be copied, or amplified, by the primers she or he chooses.
PCR is based on using the ability of DNA polymerase to synthesize new strand of DNA complementary to the offered template strand. Add 90 μL of PCR-grade water. The polymerase chain reaction (PCR, described here) works mainly because of two components – a heat-stable DNA polymerase enzyme (adds new nucleotides to a chain of nucleotides) and a pair of DNA PCR primers.
Nucleotide sequence analysis of 26 cloned PCR products showed that in PCRs with papA primers, six out of eight obtained PCR products were false due to non-specific binding of the forward primer on both. It depends on the size and whether you are sequencing a plasmid cloned DNA. In the asymmetric PCR, two primers in a ratio of 100:.
This length is long enough for adequate specificity and short enough for primers to bind easily to. Taq DNA polymerase is relatively unstable above 90°C during denaturation of DNA strands. Thermal asymmetric interlaced PCR is used to isolate unknown sequence flanking a known sequence.
Since DNA synthesis is always from 5' to 3' , the 3' ends of a PCR primer set point towards each other, when they are annealed to their template strand, and the primers anneal on opposite strands of the PCR template. Using the 8-channel pipettor, transfer 1.4 L of the reverse primer of primer set #1 to column 1 of the PCR-ready MasterMix plate. PPPP stores the gene locations and directions, along with gene synonyms, in a Perl disk-based hash (DBM or DataBase Module) file.
This is known as a GC Clamp. Here, custom-designed mutant primers oriented in the inverse direction are used to amplify the entire circular template with incorporation of the required m …. The polymerase chain reaction (PCR) uses a pair of custom primers to direct DNA elongation toward each-other at opposite ends of the sequence being amplified.
PCR amplification requires 2 primers that determine the region of sequence amplified in the forward and reverse direction. It is generally accepted that the optimal length of PCR primers is 18-22 bp. Taq polymerase works only in 5’ to 3’ direction hence the DNA synthesis occurs in the same 5’ to 3’ direction.
Criteria for PCR primer design. It attaches to the primer and then adds DNA bases to the single strand one-by-one in the 5’ to 3’ direction. PCR was invented in 1984 by the American biochemist Kary Mullis at Cetus Corporation.It is fundamental to much of genetic testing including analysis of.
In PCR, you start with many copies of each of the two primers;. The forward primer is designed along one strand in the direction toward the reverse primer. Be mindful not to have too many repeating G or C bases.
The duration of this step depends on the length of DNA sequence being amplified but usually takes around one minute to copy 1,000 DNA bases (1Kb). Each strand runs in the opposite direction to the other (ie. Although Taq DNA polymerase significantly improved PCR protocols, the enzyme still presented some drawbacks.
Unlike traditional cloning, PCR offers the ability to readily clone DNA fragments that. Nested PCR involves the use of two primer sets and two successive PCR reactions. The other forward 2 nd PCR primer contains a tag sequence, such as His-, FLAG-, or MYC-tag, therefore can differ from one another depending on your tag choice.
We synthesize our oligos in the 3' -> 5' direction off of a controlled-pore glass (CPG) solid support using amidite chemistry. PCR reaction components Master mix:. The primary benefit of Hot Start PCR is that it allows for reaction set up at room temperature without mispriming, or the formation of primer dimers for increased overall yields.
DNA polymerase extending the 3′ end of a PCR primer in the 5′ to 3′ direction. And I'm assuming since it's called Polymerase Chain Reaction, that this is where the polymerase is. You will learn in this section a detailed explanation of the PCR primer design criteria and how they affect the primer sensitivity and stability including;.
Genscript online pcr primer design tool for perfect PCR and sequencing primers design. The result is that in the next 5-10 PCR cycles, only single-stranded DNAs are generated. LUX PCR Primers These assays employ two DNA primers, one of which is.
Aliquot and store working primer solutions at - o C. PCR or polymerase chain reaction, is one of the most useful tools in a biologists tool kit. This means that the two strands are complementary to each other in the order of their bases.
Within the known sequence TAIL-PCR uses a nested pair of primers with differing annealing temperatures;. Avoid excessive freeze-thawing of. Here are some guidelines for designing your PCR primers:.
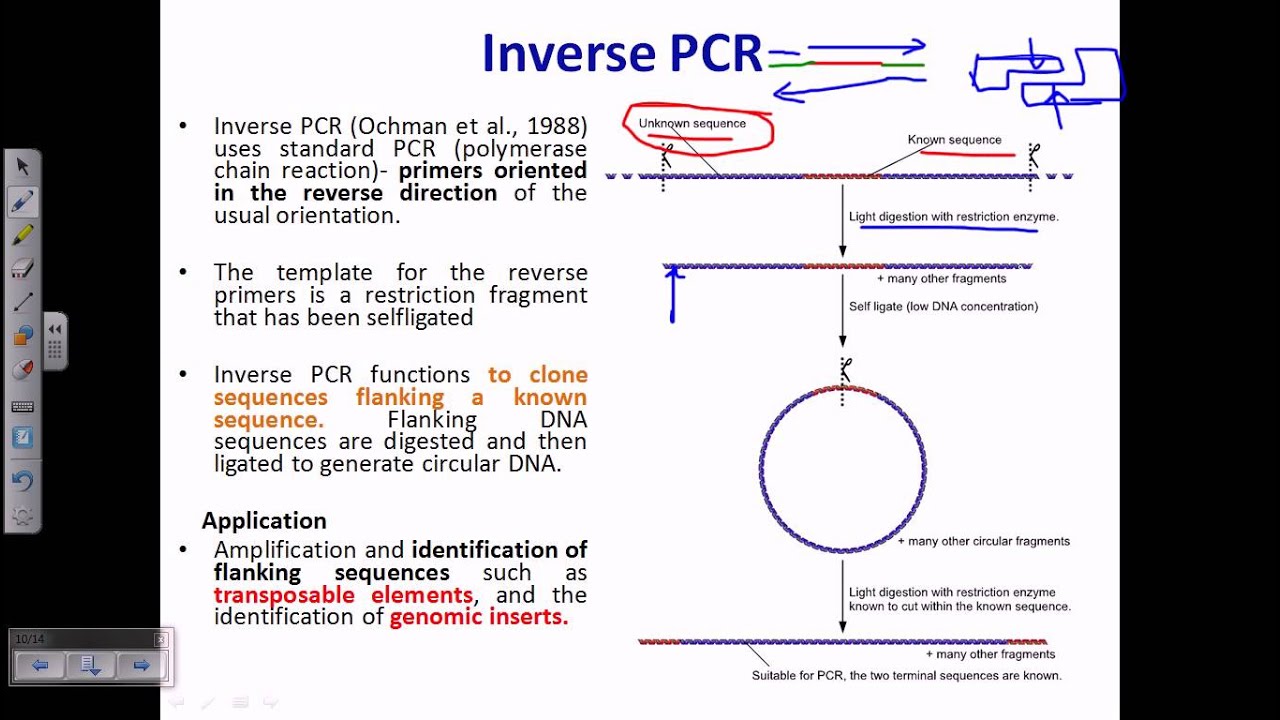
In this PCR, primers are oriented in the reverse direction of the usual orientation. Mix the MasterMix and primers in column 1 using the 8-channel pipettor. Polymerase Chain Reaction (PCR) is a technique that has various applications in research, medical, and forensic field.
These primers are typically between 18 and 24 bases in length, and must code for only the specific upstream and downstream sites of the sequence being amplified. The basic ingredients of a reaction system include a DNA template, a buffer solution, deoxyribonucleoside triphosphate (), Taq polymerase, and a pair of primers (the. When the program is run, the gene locations are used to extract sequences from the Sz.
PCR primer design. This 3' -> 5' technique mitigates the effect of truncated product on primer binding, as it will be limited to the 5' end. There are two sets of forward and reverse primer sets in the 2 nd PCR.
The template for the reverse primers is a restriction fragment that has been self-ligated Cloning of sequences flanking a known sequence. The first set of primers are designed to anneal to sequences upstream from the second set of primers and are used in an initial PCR reaction. The most common concentration for a working primer solution is 10 μM.
It amplifies the DNA fragment of interest. PCR primers Like other DNA polymerases, Taq polymerase can only make DNA if it's given a primer, a short sequence of nucleotides that provides a starting point for DNA synthesis. List of primers and probes labeled for EUA use and distributed by the International Reagent Resource may be used for viral testing with the CDC 19-nCoV Real-Time RT-PCR Diagnostic Panel.
It is also a sensitive test for disease diagnosis and genotyping. If you are unfamiliar with PCR, watch the following video:. They are anti-parallel), with only certain bases lying opposite each other (an adenine is always opposite a thymine and a guanine is always opposite a cytosine).
Scorpions PCR primers contain a sequence complementary to an internal portion of the target sequence. Additionally, they run in the 5′ to 3′ direction from left to the right. Primer length, primer melting temperature, primer annealing temperature, GC% of the primer, GC-clamp, cross homology and primer secondary structure.
This file is pre-calculated from data in Sz. Hot Start PCR enzymes are inactive at ambient temperatures and require a heat activation step for the PCR reaction to occur. They are known as forward primer and reverse primer.
For a PCR product (upto ~700 bp), if you use only the forward primer then the sequence from the start codon upto around. Aim for the GC content to be between 40 and 60% with the 3’ of a primer ending in G or C to promote binding. Repeat steps 2 an d3 for primer set #2, this time dispensing the primers into column 7 of the PCR-ready MasterMix plate.
Complentary to 3' to 5' of the target DNA Name of the primer. Hence, primers are served as starting points of the synthesis of new strands. PCR technique can also be used for the synthesis of single-stranded DNA molecules, particularly useful for DNA sequencing.
During the denaturing phase of the PCR cycle, all of the target DNA and the primers s. Forward and reverse primers are two types of primers that are useful in PCR. One must selectively block and unblock repeatedly the reactive groups on a nucleotide when adding a.
Usually only a small part of a greater whole. Occasionally, multimerization of the PCR primers can cause duplication of the primer sequence in the resulting plasmid. It allows you to take a DNA sample, select a part of it and make.
Soon there will be far more pieces of DNA that were synthesized starting with one primer thus has a definite stopping point (the DNA ends) when the synthesis starts with the other primer and goes the other way. In this lecture, I explain how to design working primers for use in PCR. It is generally accepted that the optimal length of PCR primers is 18-22 bp.
Pombe genome (Wood et. Also, their length varies between 18 to 25 base pairs. Because DNA polymerase can add a nucleotide only onto a preexisting 3'-OH group, it needs a primer to which it can add the first nucleotide.
The PCR aims to detect the presence of a section of genetic material;. The reverse and sequencing primer will be extended in 5' to 3' direction of the complement strand (in figure to the left) creating thereby a copy of the complement strand. Since DNA is double stranded, two types of primers are needed in PCR.
Oligonucleotide primers are necessary when running a PCR reaction. An additional restriction digest, which excises a short region (<400 bp) proximal to the target site, can identify these duplications (slightly larger bands sizes relative to the original template). Primer Design for PCR.
Pombe GeneDB to speed up the program for primer design without the overhead of creating a relational database. A degenerate primer is used to amplify in the other direction from the unknown sequence. To make a 10 μM working primer solution, follow these steps:.
This results in the simultaneous synthesis of two new strands of DNA:. The length of each primer should be between 18 to 25 nucleotides in length. Likewise, the reverse primer is designed from the complimentary strand.
This length is long enough for adequate specificity and short enough for primers to bind easily to the template at the annealing temperature. Each primer sequence matches a sequence on one of the two complementary strands of the target DNA. The extra long DNA synthesis will only happen with the original template DNA which goes on a long way in both directions.
For more demanding studies, such as rare variant detection, rhPrimer GEN2 designs are recommended. Is a revolutionary method developed by Kary Mullis in the 1980s. Add 10 μL of primer stock solution to an RNase- and DNAse-free tube.
The Polymerase Chain Reaction (PCR) can be used to rapidly generate DNA fragments for cloning, provided that a suitable source of template DNA exists and sufficient sequence information is known to permit design of primers specific for the desired amplicon. Polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it to a large enough amount to study in detail. In our study, we used PCR to clone papA, papEF, papG and F17G genes of Escherichia coli isolated from faecal samples of dogs with diarrhoea.
In the direction from the start codon to the stop codon from the forward primer, and in the direction from the stop codon to the start codon from the reverse primer. The PCR product size is 549 basepairs (bp), which includes the primers themselves and the bp in between the primers (segment -165). The two reverse 2 nd PCR primers and one forward 2 nd PCR primers are universal primers.
Efficient cleavage of a blocked primer by RNase H2 requires a footprint of at least 8–10 bases upstream of the single RNA base in the primer and 4 bases downstream of the RNA base. The G and C bases have stronger hydrogen bonding and help with the stability of the primer. Inverse PCR is a powerful tool for the rapid introduction of desired mutations at desired positions in a circular double-stranded DNA sequence.
Both are oligonucleotides used for the initiation of PCR. This requirement makes it possible to delineate a specific region of template sequence that the researcher wants to amplify. One needs to design primers that are complementary to the template region of DNA.
Resources and interim guidelines for laboratory professionals working with specimens from persons with coronavirus disease 19 (COVID-19).
Polymerase Chain Reaction Pcr Article Khan Academy

Inverse Polymerase Chain Reaction An Overview Sciencedirect Topics

Colony Pcr Other Means Of Plasmid Insert Investigation The Bumbling Biochemist
Pcr Primer Direction のギャラリー
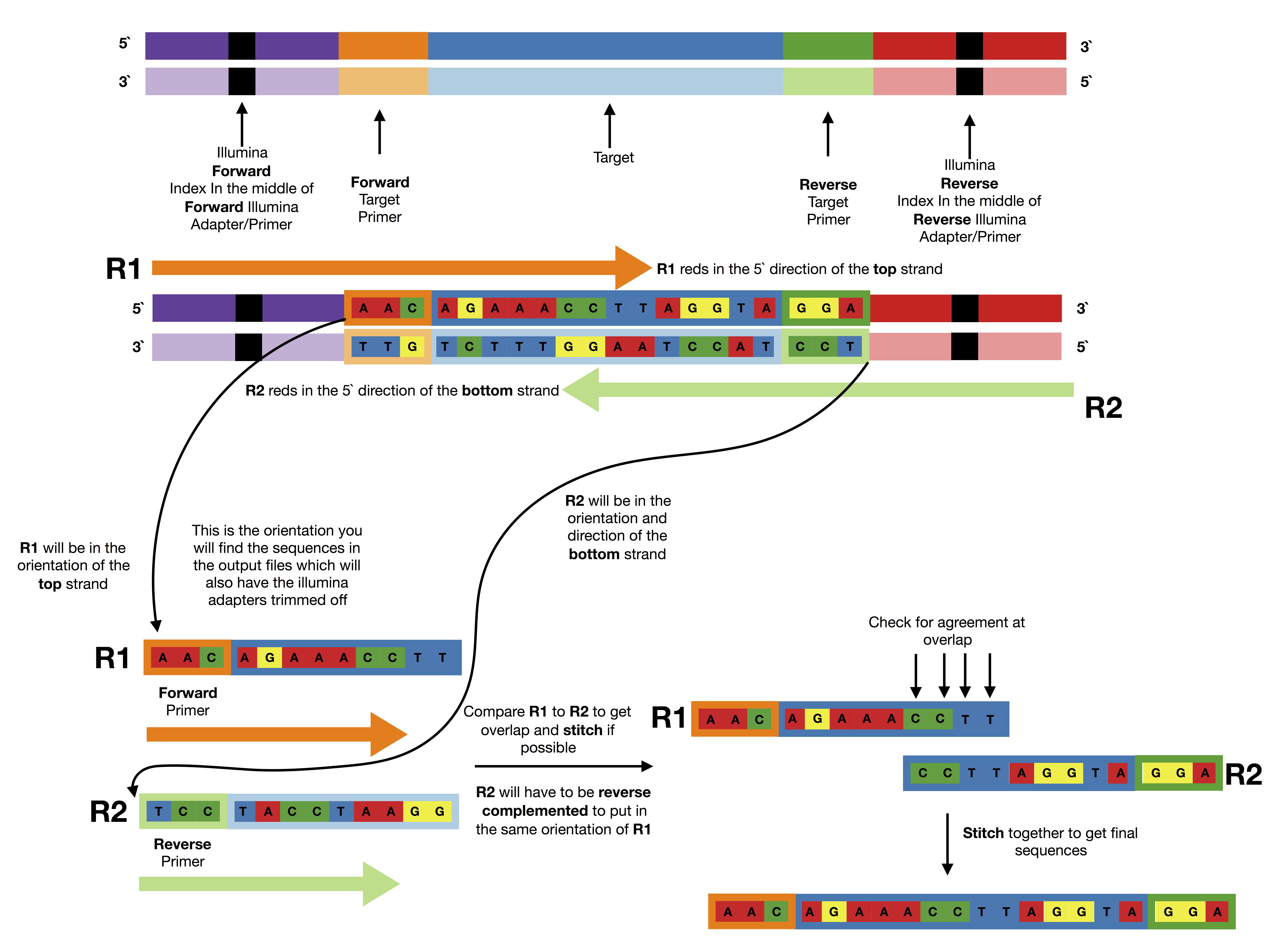
Illumina Paired End Information

Pcr Overview Goldbio

h5425 Molecular Biology And Biotechnology
What Are Forward Primer And Reverse Primer In Pcr Quora

Primer Molecular Biology Wikipedia
Polymerase Chain Reaction Pcr Article Khan Academy
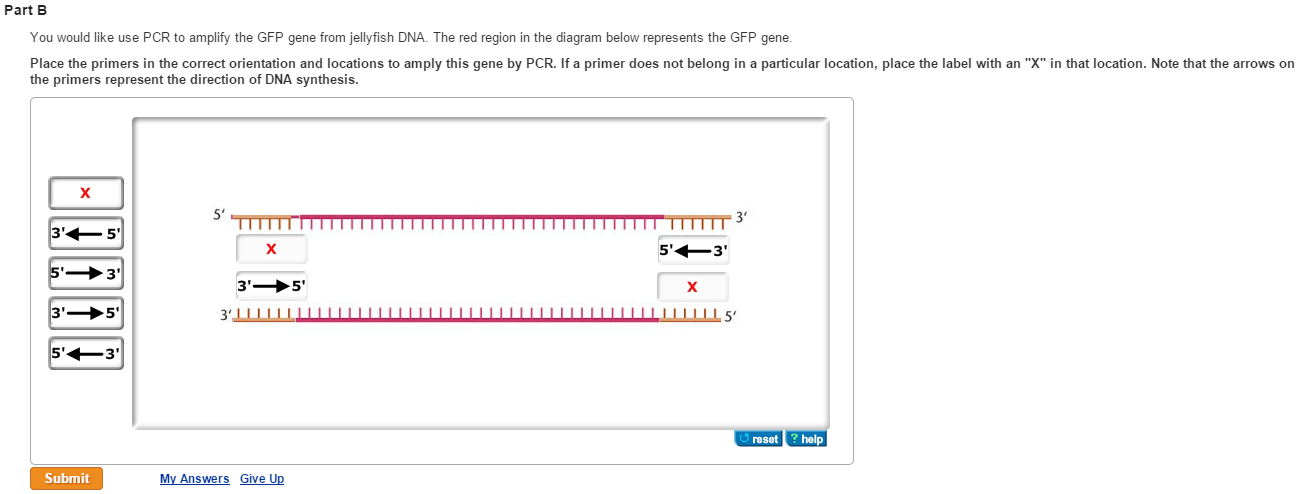
Solved You Would Like Use Pcr To Amplify The Gfp Gene Fro Chegg Com
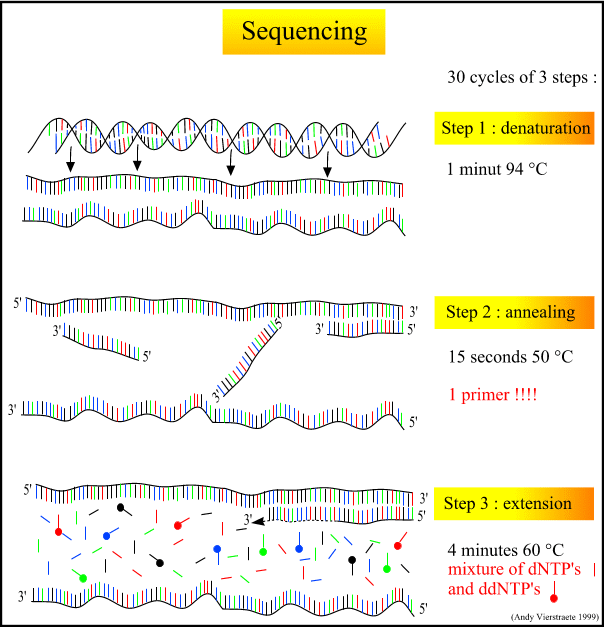
Principle Of Sequencing

Untitled Document

How To Select Primers For Polymerase Chain Reaction
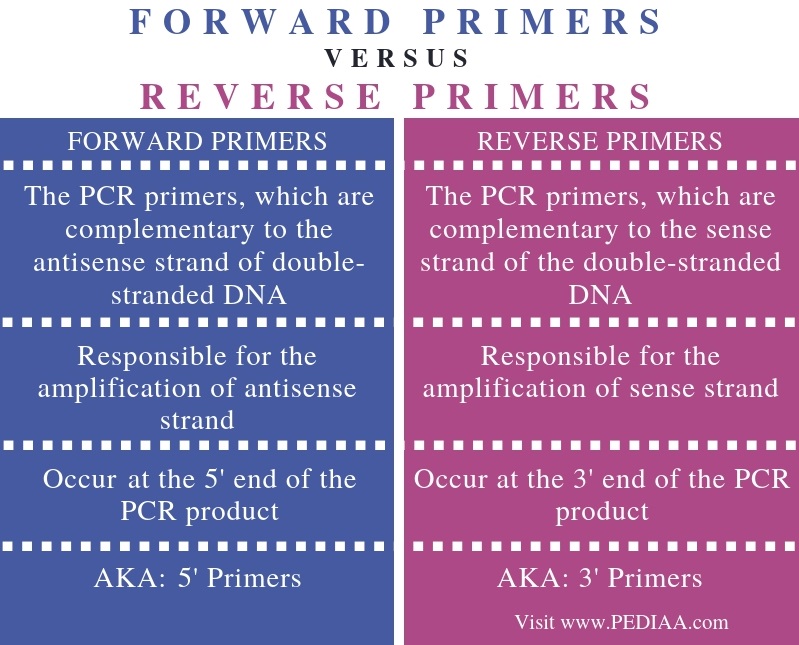
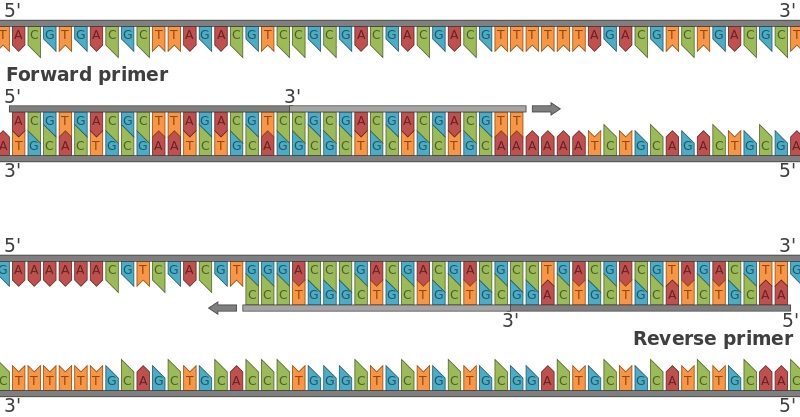
What Is The Difference Between Forward And Reverse Primers Pediaa Com

Simplified Primer Design For Pcr Based Gene Targeting And Microarray Primer Database Two Web Tools For Fission Yeast Penkett 06 Yeast Wiley Online Library

Pcr Basics Thermo Fisher Scientific Us
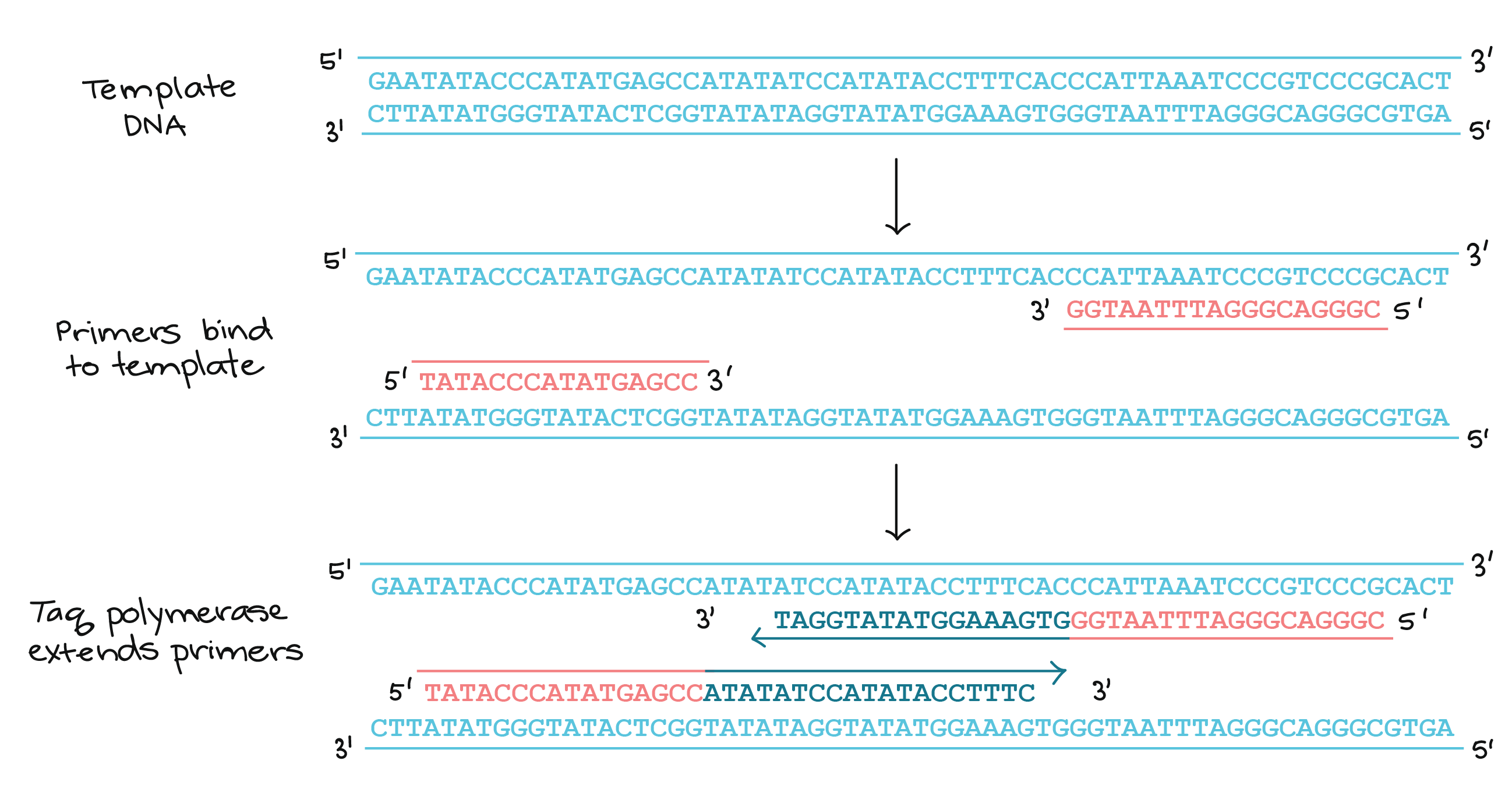
Polymerase Chain Reaction Pcr Article Khan Academy

Pcr
A Novel Universal Primer Multiplex Pcr Method With Sequencing Gel Electrophoresis Analysis

Are Primers Double Stranded Molecules If Not How Do Primer Dimers Form In A Pcr Reaction Quora
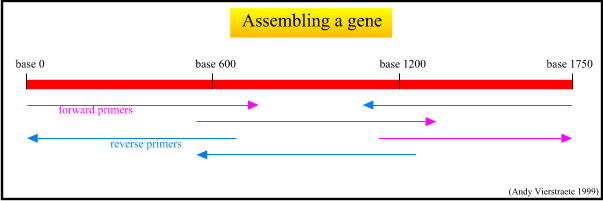
Principle Of Sequencing

Polymerase Chain Reaction Wikipedia

Primer Design Tutorial Geneious Prime
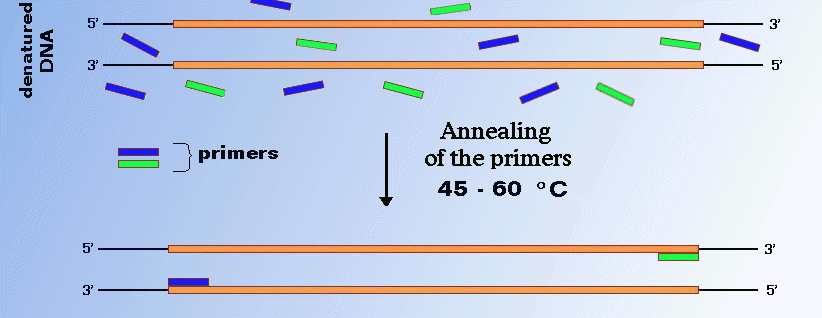
Annealing Of Primers
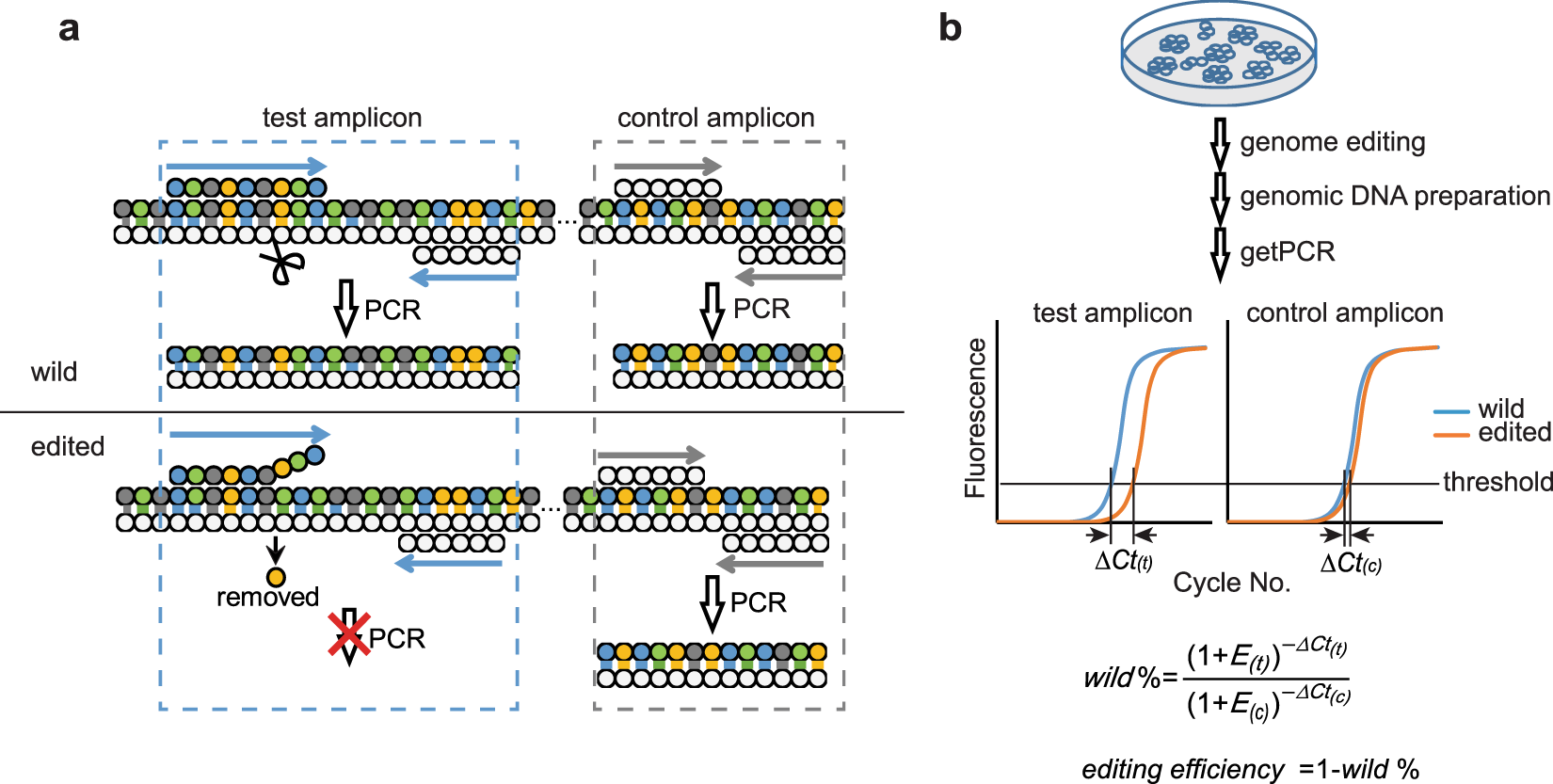
A Qpcr Method For Genome Editing Efficiency Determination And Single Cell Clone Screening In Human Cells Scientific Reports
Designing Pcr And Sanger Sequencing Primers Seq It Out 5 Behind The Bench
Solved You Would Like Use Pcr To Amplify The Gfp Gene From Jellyfish Dna The Red Region In The Diagram Below Represents The Gfp Gene Course Hero

Polymerase Chain Reaction Pcr Article Khan Academy
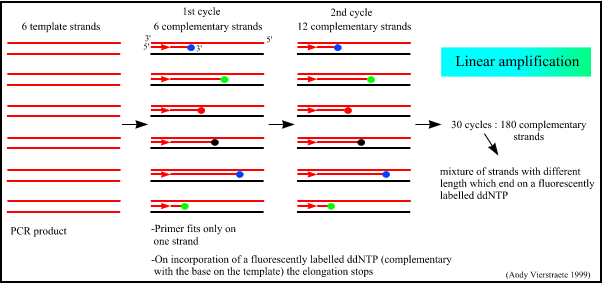
Principle Of Sequencing

Pcr Introduction Abm Inc

Pcr Overview Goldbio

Primer Molecular Biology Wikipedia
Inverse Pcr Youtube

Polymerase Chain Reaction Pcr Diamantina Institute University Of Queensland
Comparison And Validation Of Some Its Primer Pairs Useful For Fungal Metabarcoding Studies

h5425 Molecular Biology And Biotechnology

Reverse Transcription And Polymerase Chain Reaction Principles And Applications In Dentistry
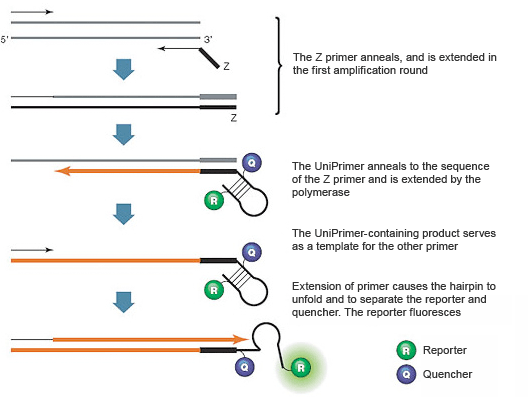
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad

Primer Design Tips For An Efficient Process
Parallel Dna Polymerase Chain Reaction Synthesis F1000research

How To Make Primers For Pcr Pediaa Com

List Of Primers Used In Quantitative Real Time Pcr Qrt Pcr Forward Download Scientific Diagram
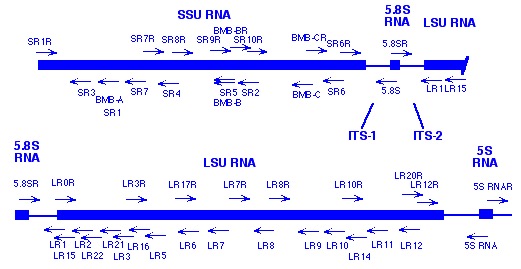
Conserved Primer Sequences For Pcr Amplification Of Fungal Rdna Vilgalys Mycology Lab Duke University
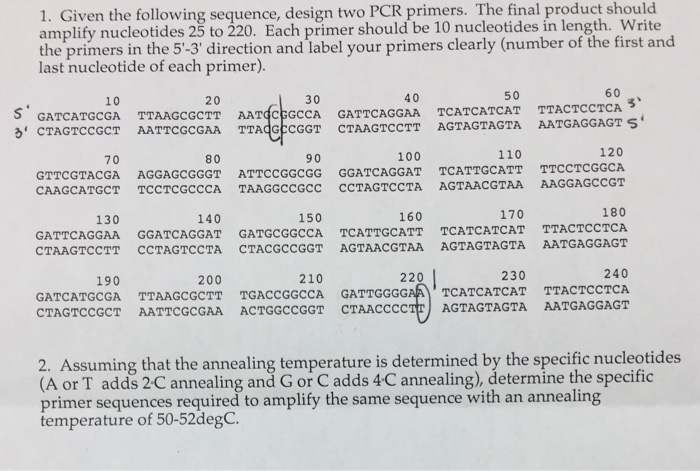
Solved Given The Following Sequence Design Two Pcr Prime Chegg Com

Pcr And Molecular Biology Fundamental Principles
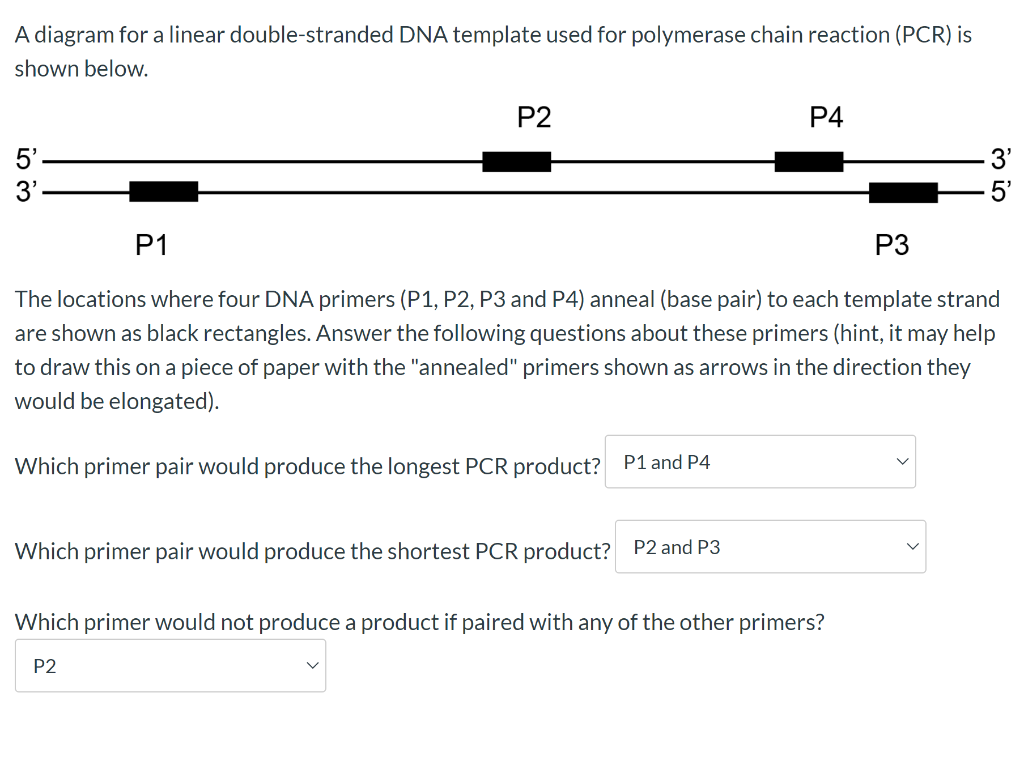
Solved The Locations Where Four Dna Primers P1 P2 P3 A Chegg Com

Merritt Genomics

Schematic Representations Of The Pcr Primer Positions For Multiplex Pcr Download Scientific Diagram
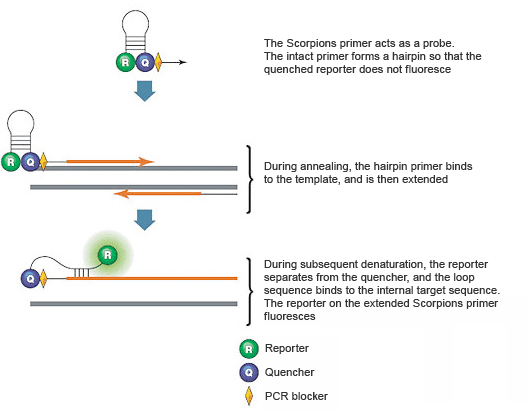
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad

Inverse Polymerase Chain Reaction An Overview Sciencedirect Topics

Solved 2 A Design A Pair Of Pcr Primers That Could Pote Chegg Com

Polymerase Chain Reaction Pcr Diamantina Institute University Of Queensland
Plos One Allele Specific Dna Methylation Of Disease Susceptibility Genes In Japanese Patients With Inflammatory Bowel Disease
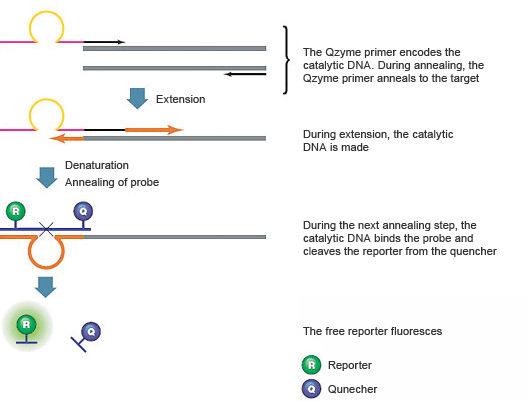
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad
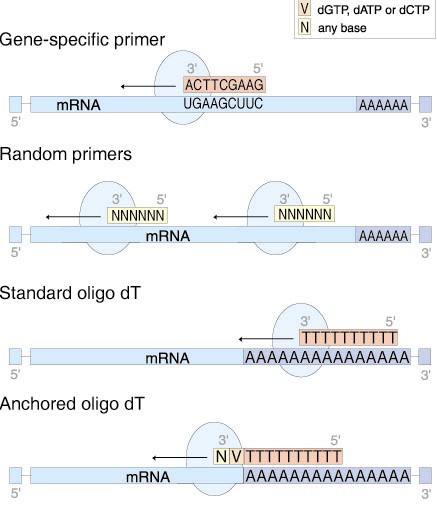
Www Gene Quantification Info

Pcr Primers Used For Sanger Sequencing Gene Direction Sequence 5 To Download Scientific Diagram

Inverse Pcr Youtube
Q Tbn 3aand9gcqksw6z5yfg9k6y7nfzbgoh65zaj7edursq Hmswh5zid18pniz Usqp Cau

Colony Pcr Other Means Of Plasmid Insert Investigation The Bumbling Biochemist

Pcr Overview Goldbio

Sequences Of Real Time Pcr Primers Sybr Green Primers Download Table
Q Tbn 3aand9gcqzninymvxcoxl6ityd4zga 5jbhu67honfuwvj3pcas6qidmae Usqp Cau
Primer Design
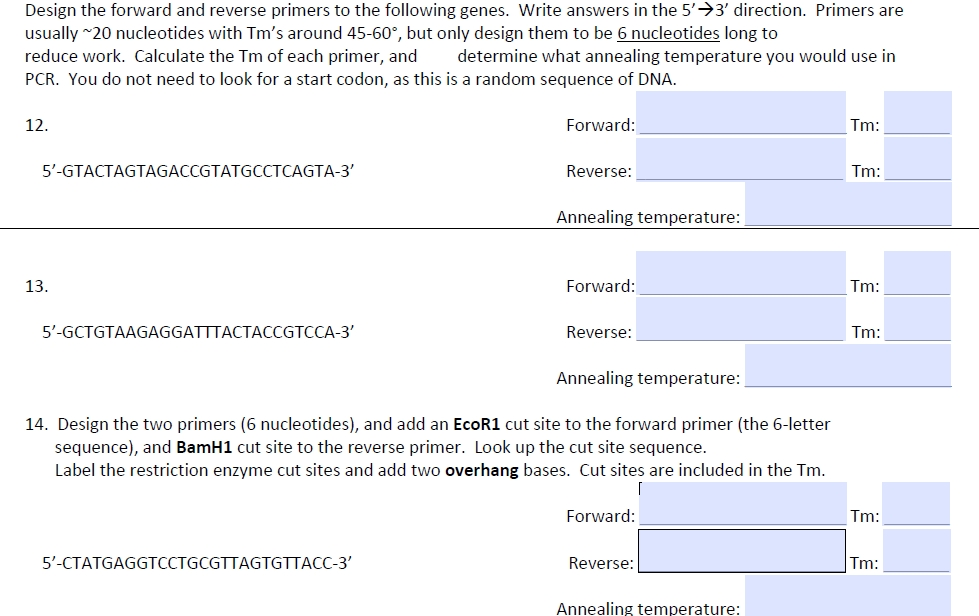
Solved Design The Forward And Reverse Primers To The Foll Chegg Com

Biochemistry Class Notes Polymerase Chain Reaction
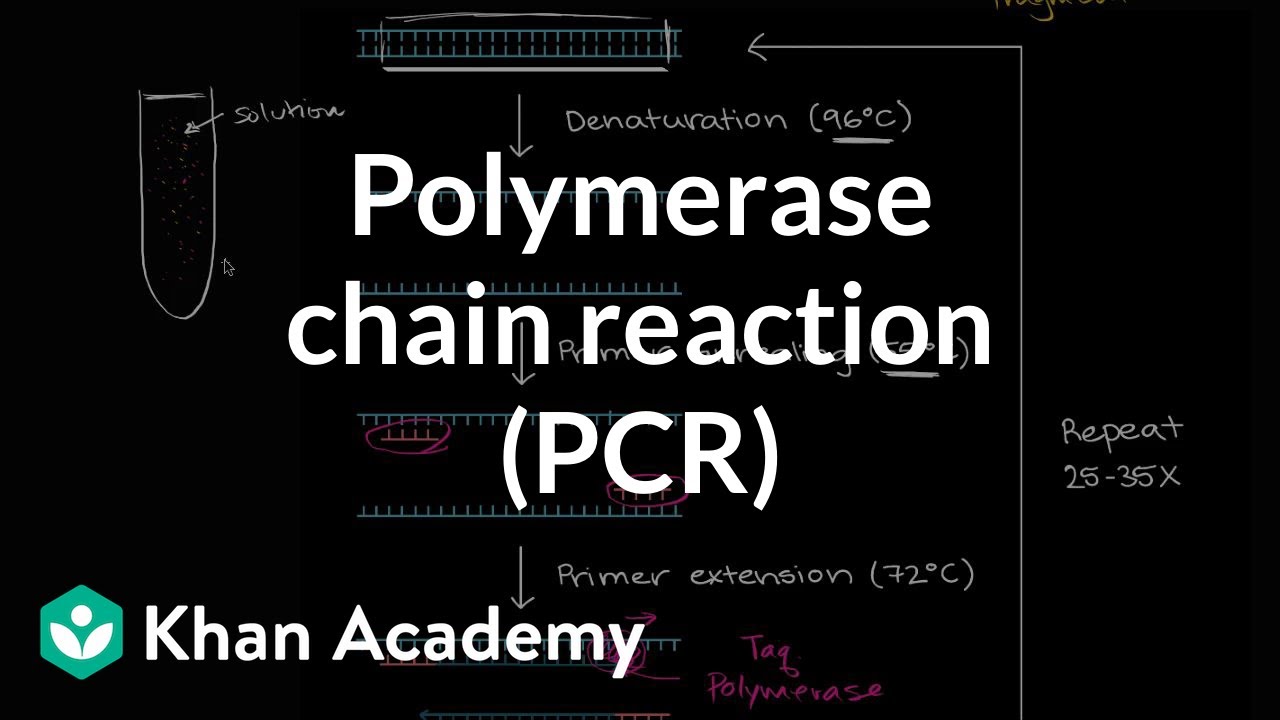
Polymerase Chain Reaction Pcr Video Khan Academy

Primer Sequences For Pcr Analysis Of Mer Loci Mer Primers A Direction B Download Table

Dna Walking Strategy A Designed Primer Position Of T35s Pcambia A R Download Scientific Diagram
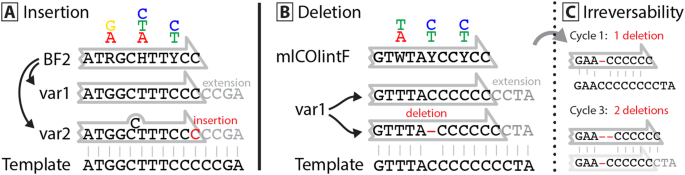
Slippage Of Degenerate Primers Can Cause Variation In Amplicon Length Scientific Reports
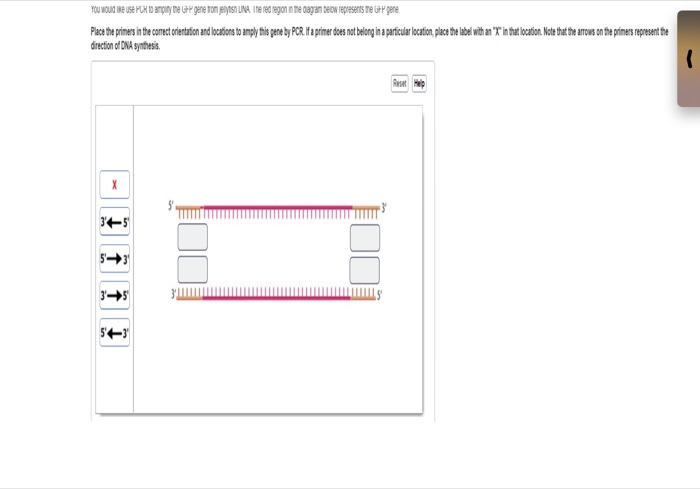
Solved Pace The Priners In The Comect Orientation And Loc Chegg Com
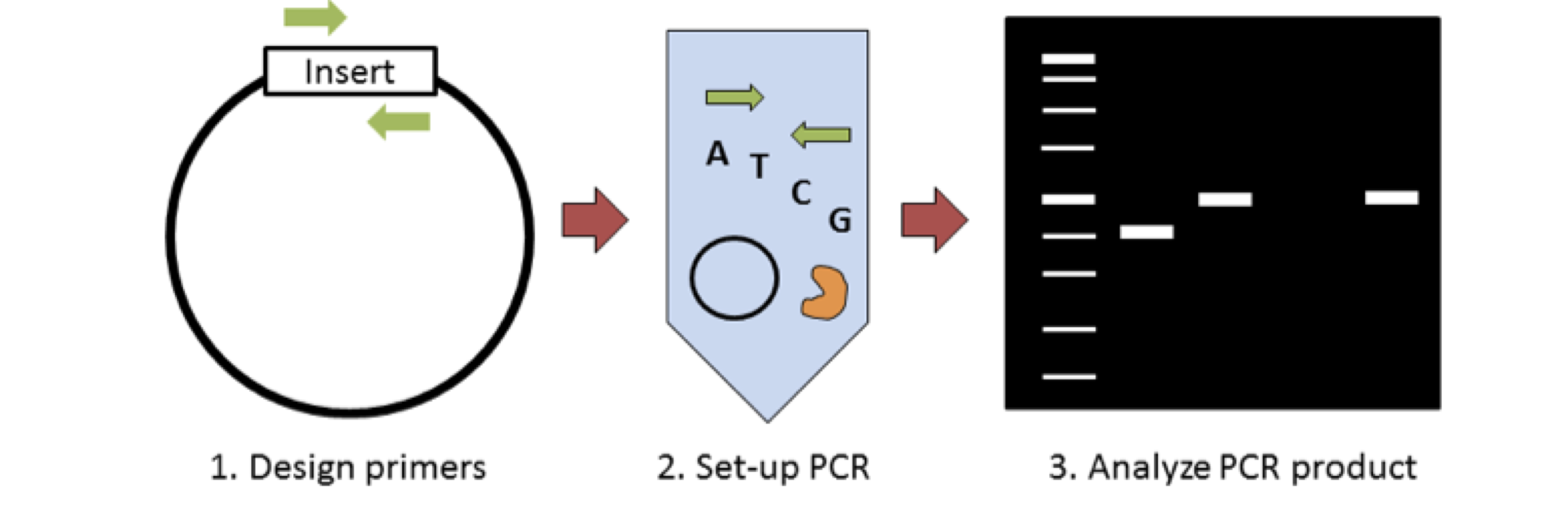
Plasmids 101 Colony Pcr
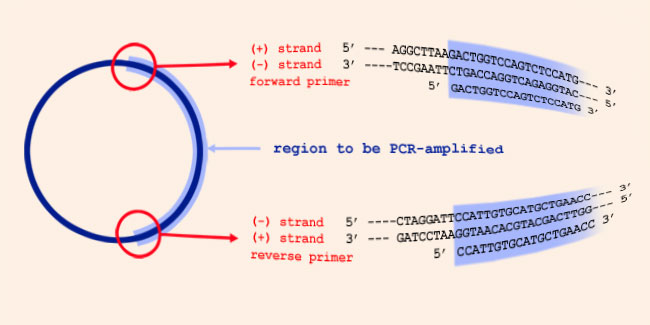
Sequence Notation
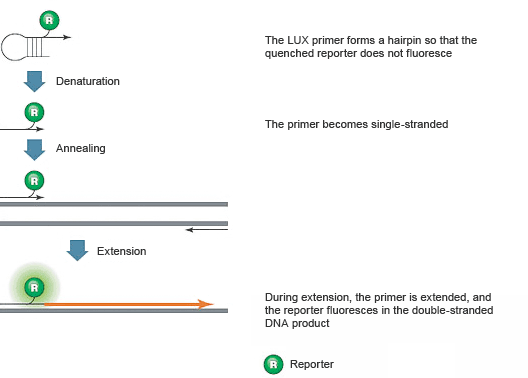
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad
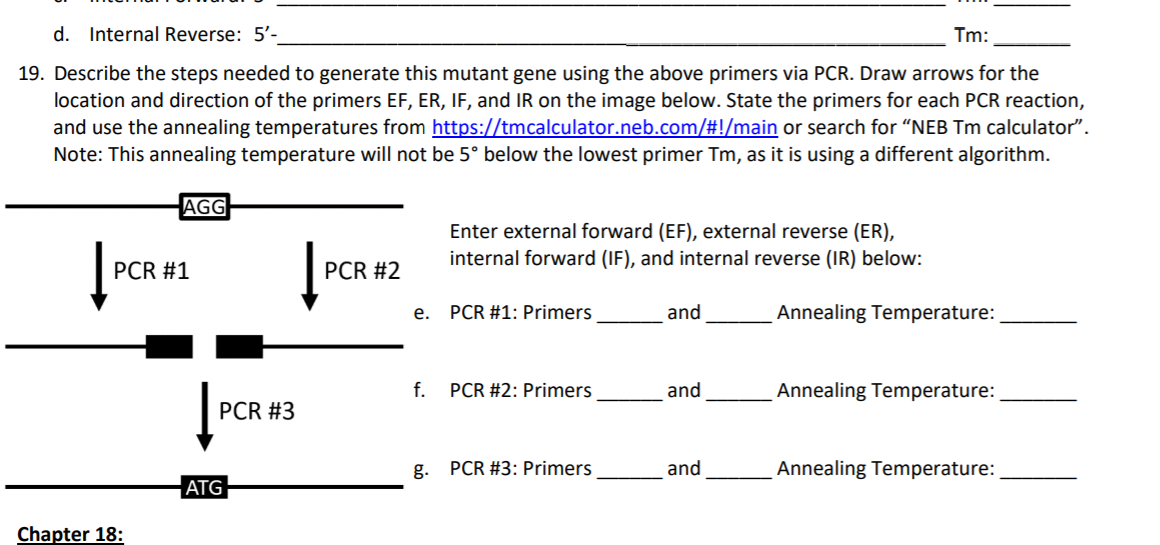
D Internal Reverse 5 Tm 19 Describe The Step Chegg Com

Pcr Primer Sequences In 59 39 Direction Download Table

Pcr Primers A Primer Virology Down Under
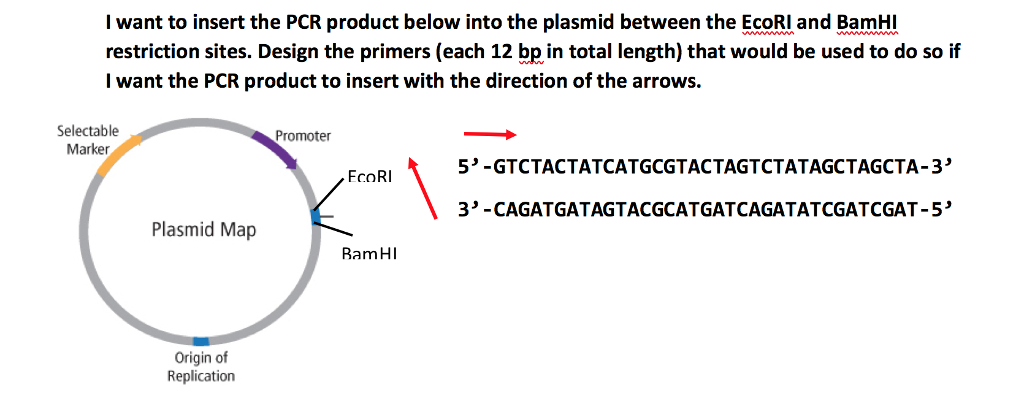
Solved I Want To Insert The Pcr Product Below Into The Pl Chegg Com

Addgene What Is Polymerase Chain Reaction Pcr

Addgene Protocol How To Design Primers
Q Tbn 3aand9gcqksw6z5yfg9k6y7nfzbgoh65zaj7edursq Hmswh5zid18pniz Usqp Cau

Addgene Plasmid Cloning By Pcr With Protocols

Primer Selection Guidelines Good Primers Important For Pcr And Automated Sequencing Methods And Technology For Genetic Analysis

Solved Design 15 Nucleotide Long Pcr Primers From The Beg Chegg Com

Primer Design Tutorial Geneious Prime

Primers Used In This Study Their Usage Direction Sequence And Pcr Download Table

Pcr Overview Goldbio

Untitled Document

Pcr Overview Goldbio

Pcr Primer Sequences Primers Are Shown In The 5 To 3 Direction Download Table
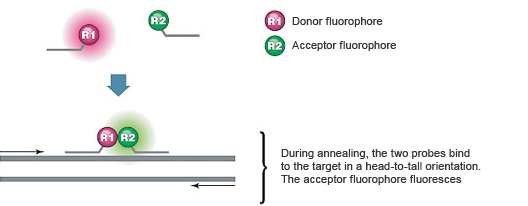
Introduction To Pcr Primer Probe Chemistries Lsr Bio Rad

Pcr Overview Goldbio
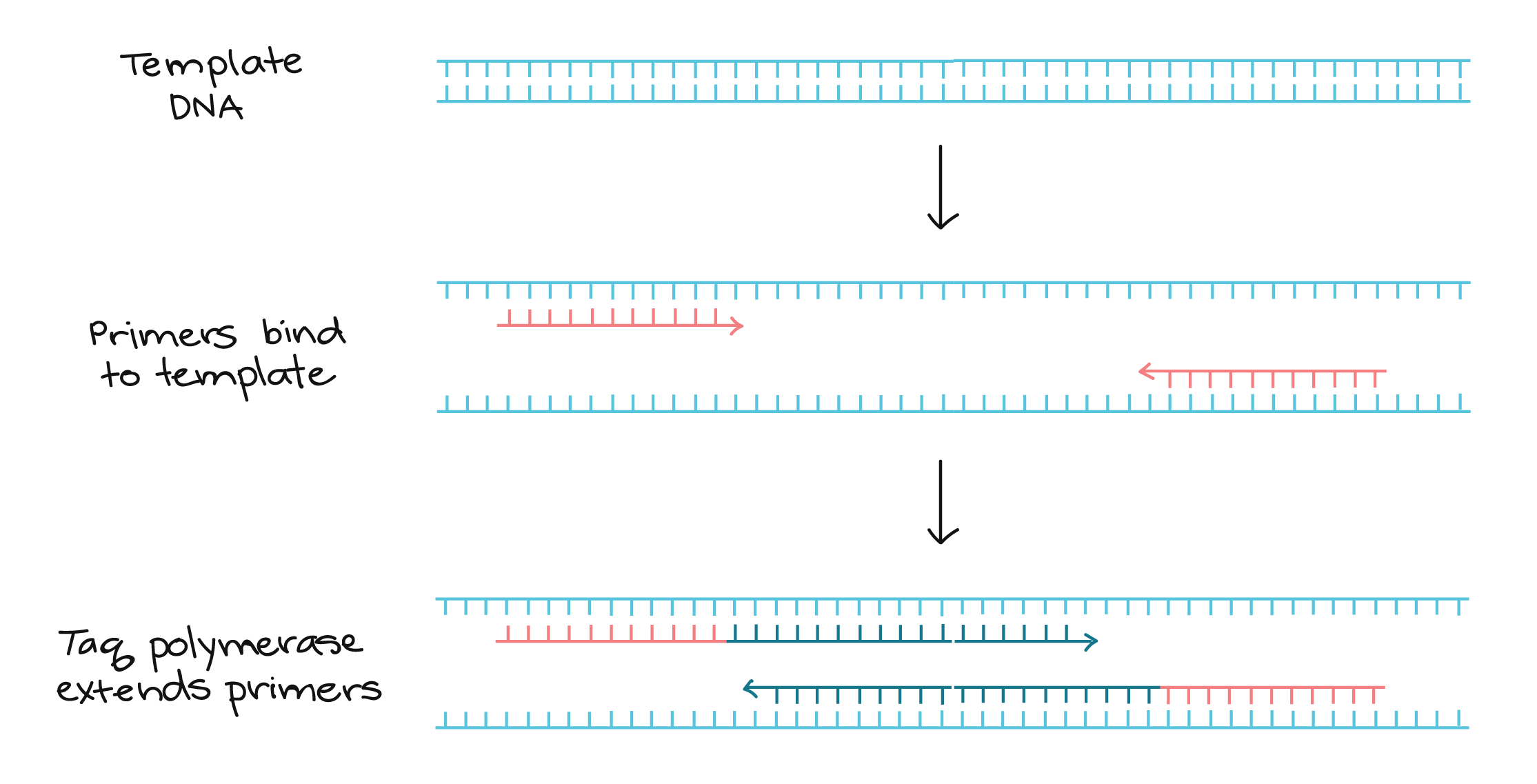
Polymerase Chain Reaction Pcr Article Khan Academy

Pcr Overview Goldbio

Merritt Genomics

Solved Part B You Would Like Use Pcr To Amplify The Gfp G Chegg Com
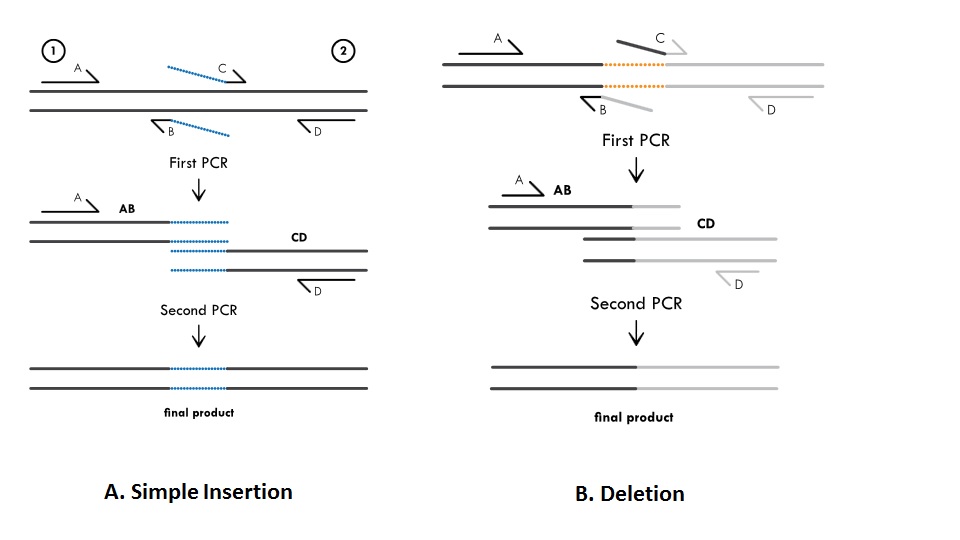
Methods For Site Directed Mutagenesis
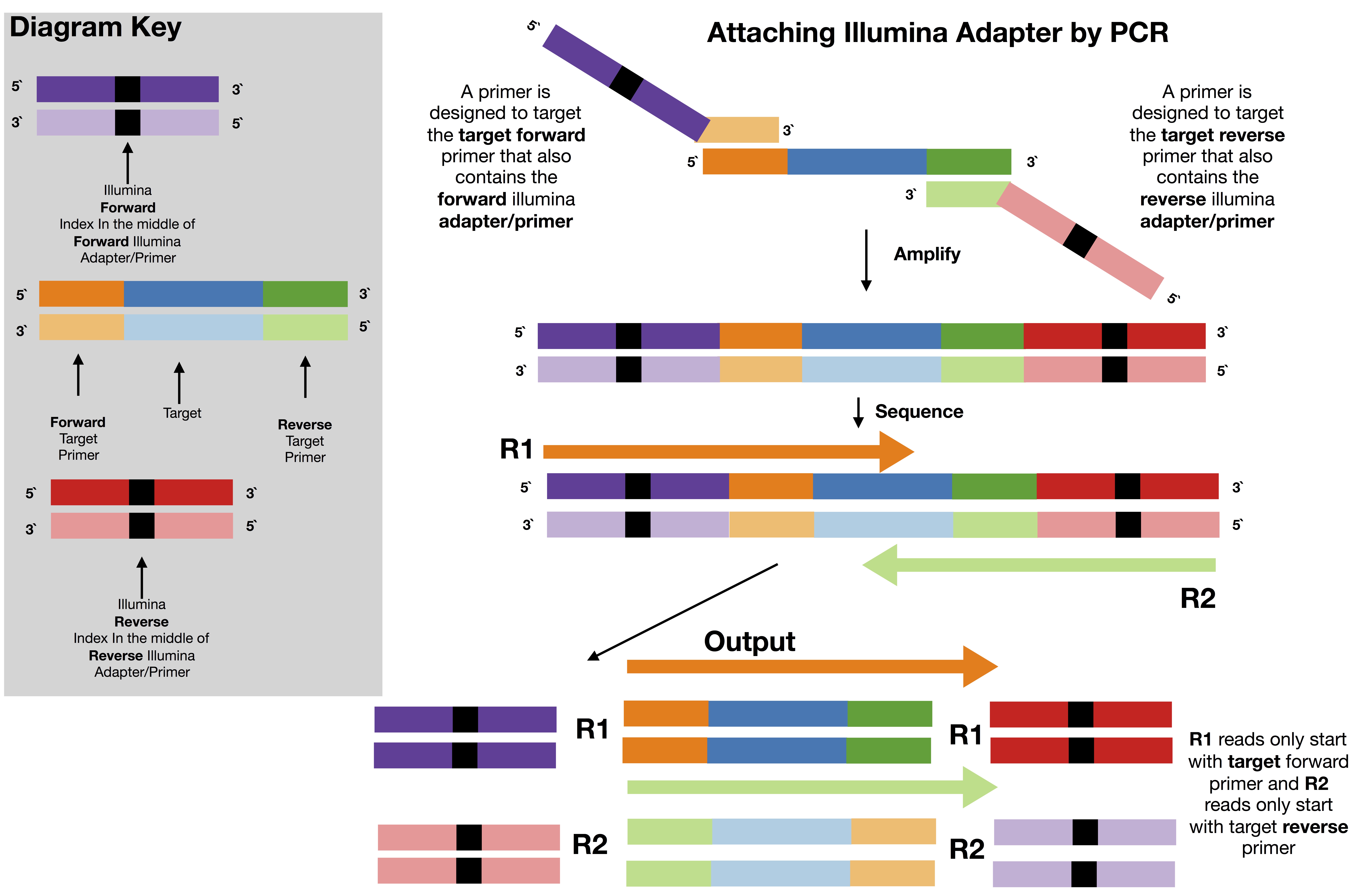
Illumina Paired End Information

A Maps Of Plasmids And Pcr Primers Used In This Report Arrows Point Download Scientific Diagram

Primer Design Tutorial Geneious Prime
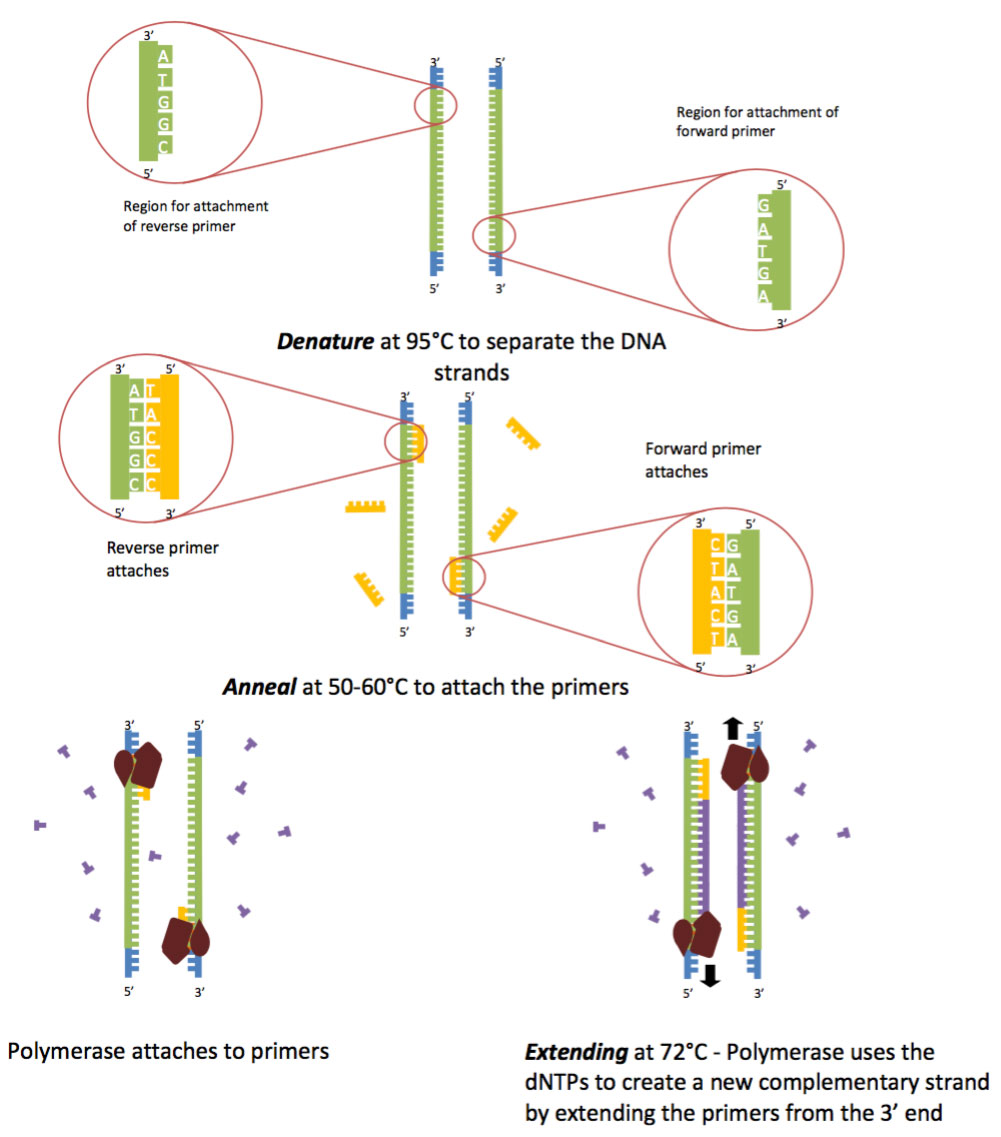
Polymerase Chain Reaction Pcr Diamantina Institute University Of Queensland
Q Tbn 3aand9gctvfxchhuftpc Zex Bfuwfrm9 C8 Dg3mp9u1pmqfbzgxvz Go Usqp Cau
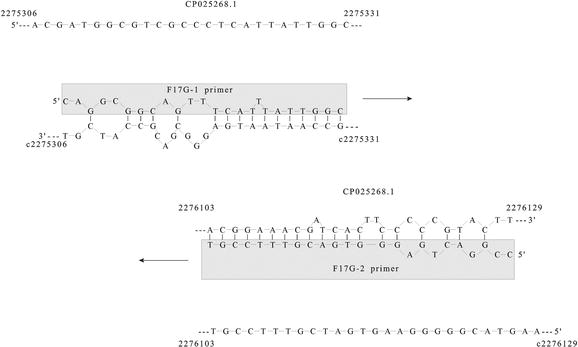
Annealing Temperature Of 55 C And Specificity Of Primer Binding In Pcr Reactions Intechopen

Primer Walking An Overview Sciencedirect Topics
What Is The Difference Between Forward And Reverse Primers Pediaa Com



