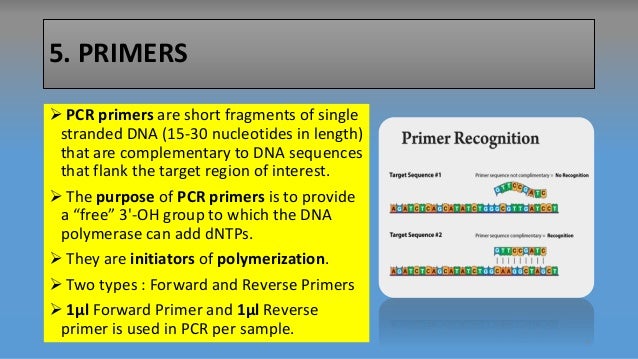
Pcr Primers Forward And Reverse
Increase Mg 2+ ion concentration generally increases PCR.

Pcr primers forward and reverse. Real-time PCR is performed using a miScript Primer Assay (forward primer) and the miScript SYBR Green PCR Kit, which contains the miScript Universal Primer (reverse primer) and QuantiTect SYBR Green PCR Master Mix. The polymerase chain reaction (PCR) uses a pair of custom primers to direct DNA elongation toward each-other at opposite ends of the sequence being amplified. Genomic DNA is introduced into a reaction mixture consisting of TaqMan® Genotyping Master Mix, forward and reverse primers and two TaqMan® MGB Probes.
19-Novel Coronavirus (19-nCoV) Real-time rRT-PCR Panel Primers and Probes;. General Rules for Designing PCR Primers, Xu lab General Rules of Primer Design for Restriction enzyme cloning A. PCR is exponential amplification in which the newly generated PCR fragment from one cycle also acts as a template for the next cycle (Figure 1A).
DNA taq polymerase Catalyze DNA amplification. The reverse primer is designed to attach to the complementary strand to synthesize DNA in the reverse direction — towards the forward primer. The forward primer will anneal with 3’-5’ DNA strand and the reverse primer will anneal with the 5´-3’ DNA strand.
The reverse primer to position is 1787. SHOW YOUR MATHEMATICAL WORK BELOW TO DETERMINE:. If we consider the sense strand (5’-3’) of a gene, for designing primers, then forward primer is the beginning of the gene and the reverse primer is the reverse-compliment.
Nearly all “experts” in PCR design would claim to believe in myth 3. The primers are added to PCR experiments to initiate the process of replication by providing the initial nucleotides to the new strand. This is a very basic guide.
If a miScript Primer Assay for the small RNA of interest is not already available, one can be custom-designed. As I'm doing qPCR almost on a daily basis, I am wondering if there is any reason not to prepare a master mix of H2O, forward primer and reverse primer in one. 2 Primers (forward and reverse) to start the process of replication.
The forward primer is designed along one strand in the direction toward the reverse primer. A master mix of all components (except template) necessary for PCR contains what basic ingredients?. The amount of 2X master-mix that is to be placed into each reaction C.
If you are unfamiliar with PCR, watch the following video:. Select the 3’ bases for the reverse primer ensuring that these are not complimentary to the forward primer. You will immediately notice, that by making the selection conditions for the primers more stringent, the list of possible primers will diminish.
Primer Design for Restriction Enzyme Cloning (E6901) Protocols.io also provides an interactive version of this protocol where you can discover and share optimizations with the research community. Likewise, the reverse primer is designed from the complimentary strand. Both Forward and Reverse primers usually consist.
Basically, I'm trying to save time:. Input the PCR template information into Primer-BLAST. There are two sets of forward and reverse primer sets in the 2 nd PCR.
Two oligonucleotide primers (one is a reverse primer and other is a forward primer) bind to target sequences in preparation for directed DNA synthesis by Taq What happens during extension?. Design forward and reverse primers for a PCR, given the target sequence. How to Make Primers for PCR The direction of both forward and reverse primer should be 5′ to 3′.
Lux AB Primers 5’ CACC ATG AAGTTTGGAAATATTTG 3’ (Forward Primer) 3’ TACTTCAAACCTTTATAAAC 5’ (Reverse Primer) 3’ TTTTAGCTTTACTTAAATGG 5’ 5’ AAAATCGAAATGAATTTACC 3’ Forward Primer = nucleotides 4230-4249 in template (+ 4 additional nucleotides) Reverse Primer = nucleotides 6290-6310 in template Total length PCR product = 80 base. The set of primers should flank the fragment you intend to amplify from the DNA template. GAC CCC AAA ATC AGC GAA AT:.
If the sample is RNA, RNA must be converted to cDNA by reverse. This is similar to using Reverse e-PCR to search a sequence database with STS markers. PCR, Forward and Reverse Primers Polymerase Chain Reaction, PCR is used to amplify a segment of DNA or RNA exponentially.
What are the Similarities Between Forward and Reverse Primer?. During Polymerase Chain Reaction (PCR) the primers will be extended from the 3'-end (-->). The forward primer attaches to the start codon of the template DNA (the anti-sense strand), while the reverse primer attaches to the stop codon of the complementary strand of DNA (the sense strand).
Primer concentration is one variable dependent on the total volume of the PCR reaction in order that sufficient copies of the primer find the target annealing sites. This is similar to using Forward e-PCR to locate STS markers in the provided sequence. Just for getting your feet w.
The 5' ends of both primers bind to the 3' end of each DNA strand. TCT GGT TAC TGC CAG TTG AAT CTG:. If you want to do a PCR, you need to enhance both strands, so you need a primer for one strand, called the forward primer, which is the beginning of your gene, and an other primer that will begin.
Buffer and MgCl2 Maintains desirable pH. Using wild-type DNA, PCR was run with both wild-type primers and the mutant forward primer with its reverse primer. Check the specificity of designed, or supplied, primers.
Nothing was observed in lane 1 using the mutant forward primer and its reverse primer, because there is a mismatch at the 3’ end of mutant. Mg 2+ ion is a cofactor for taq polymerase. Two primers are used in PCR as forward and reverse to replicate both strands of the sample DNA.
Otherwise, instead of binding with target sequences, both primer will bind with each other and creates a dimer. This is called the “reverse complement” of the top strand. PCR primers direction RE recognition sites notation Reverse complement.
Excessive concentrations of forward and reverse primers can also cause formation of primer dimer when the primers anneal and amplify themselves independent of the target DNA. Designing Forward and Reverse Primers to Have Matching Tm’s Is the Best Strategy to Optimize for PCR. Add 15×T to the primer (e.g., 5’ T 15 N 5 3’).
Both Forward and Reverse primers are made from oligonucleotides. In this lecture, I explain how to design working primers for use in PCR. The other forward 2 nd PCR primer contains a tag sequence, such as His-, FLAG-, or MYC-tag, therefore can differ from one another depending on your tag choice.
These primers are typically between 18 and 24 bases in length, and must code for only the specific upstream and downstream sites of the sequence being amplified. Flag for information on PCR products (P, present;. So, primer sequence atgcgtccggcgtagag means 5’ atgcgtccggcgtagag 3’ and the direction of the sequence of plasmid vector pGT4 is in the following orientation:.
The GC content of primers be between 40 and 60% and the presence of a C or G in the 3’end of the primer may promote. Primer design is a critical step in a PCR protocol. ~30 nucleotides in length, ideally;.
Name Description Oligonucleotide Sequence (5’>3’) Label 1 Final Conc. These circumstances can lead to self-dimers or primer-dimers instead of annealing to the desired DNA sequences. Assess the terminal 5 bases as described for the forward primer.
Appropriate restriction sites, absent in the target gene, are incorporated in the forward and reverse primers when a target gene is generated by PCR. Complementation in forward and reverse primers:. The two reverse 2 nd PCR primers and one forward 2 nd PCR primers are universal primers.
Each TaqMan MGB Probe anneals specifically to a complementary sequence, if present, between the forward and reverse primer sites. The total volume of all of the reactions b. Since you know the sequence which you want to amplify already then, it is easy to design forward and reverse primers.
Afterwards, you should design two primers, the forward and the reverse primer. Primer direction relative to the gene (forward and reverse primers are given on separate lines). Both forward primer and reverse primer should be in 5’ to 3’ direction;.
A few primers have been designed with an old version of the genome and were either incorrect or better primers have been designed;. Analyze the T m of the forward primer using a nearest neighbor algorithm. In these cases, the old primers are.
Primers should contain the complementary nucleotides to the flanking end of the DNA that wants to amplify. The fi nal concentration of Taq polymerase is to be 0.01 units/ μ L in a 50- μ L PCR. The forward primers need to bind to the 3’ end of the bottom strand and so is identical to the top strand!.
DNA polymerase extends the primers using dNTPs including the reaction. To verify correct insertion use WT Check primers, or 5UTR / 3UTR Check primers. Standard notation of DNA sequences is from 5’ to 3’.
40~60% GC content, ideally D. All four dNTPs (dATP, dCTP, dGTP, dTTP) Nucleotides (building blocks) required to construct the replicated DNA. The forward primer from position is 630.
Forward primers refer to the PCR primers, which are complementary to the antisense strand of double-stranded. The above two parameters confines the range within the CDS region of p53-RC. See the figure below,.
The reverse complement of 40bp downstream to the gene’s Stop-codon (including the Stop-codon in the primer), followed by the “reverse primer” sequence of the transformation cassette (plasmid dependent). Because primers are read and created by humans our reverse primer need to be written from the beginning to the end. Moreover, forward primers are responsible for the amplification of the antisense strand, while reverse.
Most current software packages base their design strategy on this myth. That means our hypothetical forward primer would be ATGA. These primers are designed to be complementary to the nucleotide sequences at the beginning and the end of the section of DNA we want to amplify Buffers and salts to create the correct conditions for the enzyme to function A lot of work has to go into designing the primers.
OMelting temperature should be within 55 C to 65oC. In the example, the RefSeq accession is NM_.1. Forward and reverse primers.
Some careful thought, however, quickly reveals the deficiencies of that approach. Avoid intra-primer homology (more than 3 bases that complement within the primer) or inter-primer homology (forward and reverse primers having complementary sequences). While designing primer, keep in mind that both forward and reverse primer do not match with each other or are not complementary with each other.
Basic concept of how to design forward and reverse primers for polymerase chain reaction (PCR) NOTE:. 2.Diff erentiate between target amplifi cation and signal amplifi cation. It uses the latest version of the primer3 program, and allows adjustment of most of the same primer3 parameters.
In the first round of PCR, it is the forward primer that first binds to the template, after the first round reaction, a complementary strand of DNA is synthesized and then the reverse primer binds to it and initiate the synthesize of additional strands of DNA. The forward primer is complimentary to the 3’ end of antisense strand (3’-5’) and the reverse primer is complimentary to the 3’ end of sense strand (5’-3’). Note that the position range of forward primer may not overlap with that of reverse primer.
You should go genebank sequences and browse for your gene, in this way you. The grids show all the possible forward (left side) and reverse (right side) primers that conform to the set parameters, and fall within the specified target regions for each primer. Both Forward and Reverse Primers possess short nucleotide sequence complementary to the flanking ends of the DNA double.
Primers should have melting temperature between 55 – 65 0 C;. In figure 3, two different annealing temperatures of 46°C and 55°C were employed. Identify the initiation points of PCR amplification.
G and C content should be between 50 to 60%. Assume the forward and reverse primers that you are using are in a 10uM concentration and the final concentration of PCR primers is 0.4 um. The forward primer will be extended in 5' to 3' direction.
The reverse and sequencing primer's sequences ('Right Primer') are identical to the complement sequence and bind therefore on the reference strand (shown positioned above reference strand). The length of each primer should be between 18 to 25 nucleotides in length.
How Can I Trim And Assemble My Forward And Reverse Sanger Sequence For Each Sample In Batch Using Qiagen Clc Genomics Workbench

Real Time Rt Pcr Primers Forward And Reverse Sequences Download Table
h5425 Molecular Biology And Biotechnology
Pcr Primers Forward And Reverse のギャラリー
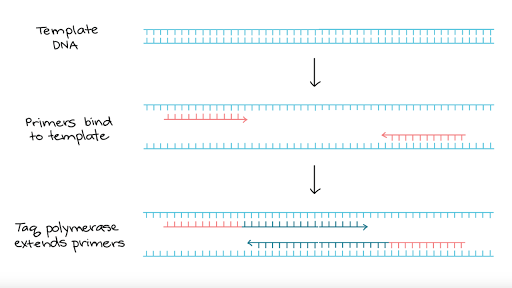
Polymerase Chain Reaction Pcr Article Khan Academy
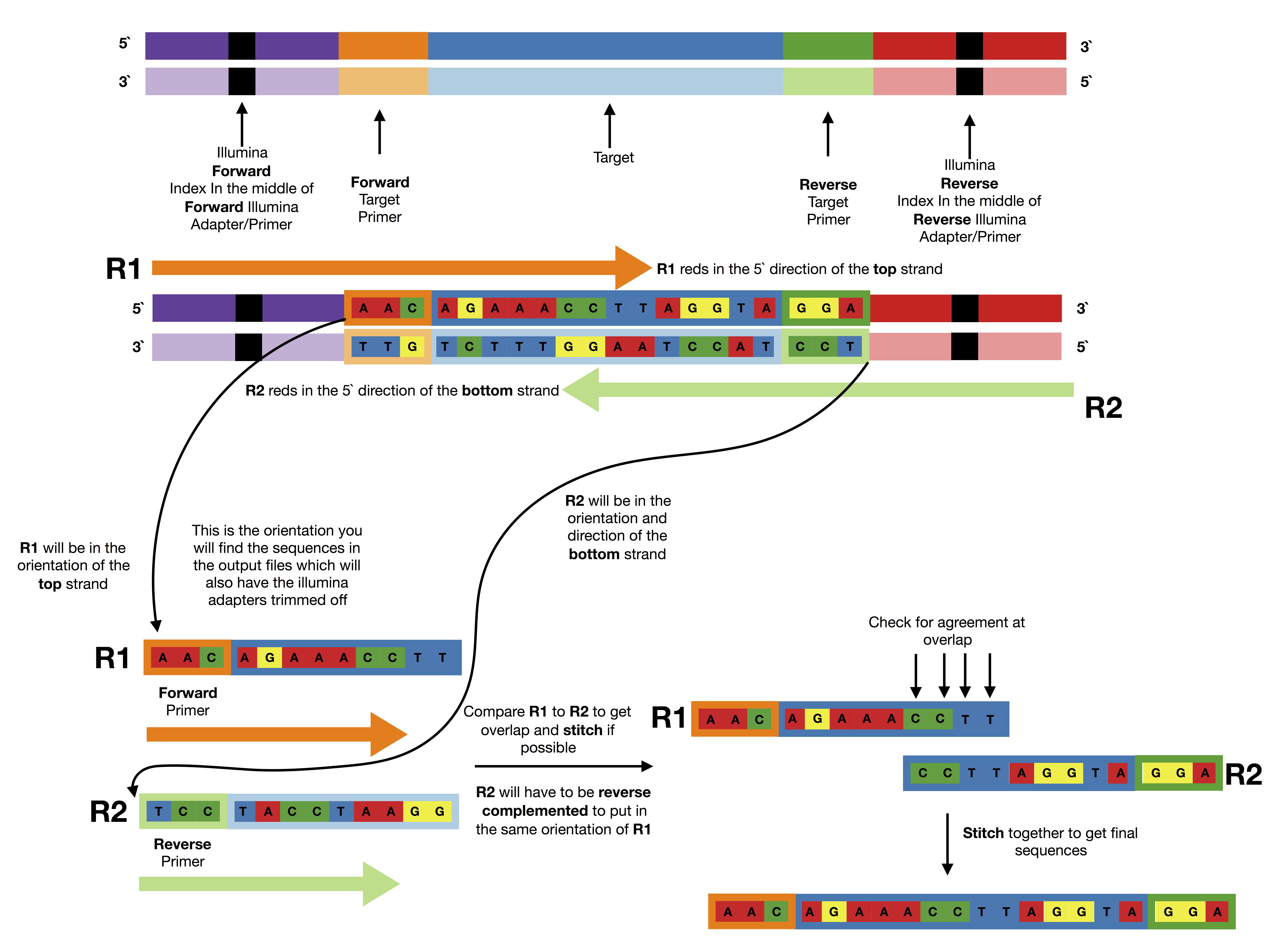
Illumina Paired End Information

Forward And Reverse Sequences Of Rt Pcr Primers Download Table

Adapter Design Pcr Amplification Of Fragments

Rt Pcr Forward And Reverse Primer Sequences Download Table

Supplementary Table 1 Forward And Reverse Primers Used In Pcr

Primer Design Tool For The 1st Pcr And Instruction Of How To Use This Tool Sigma Aldrich
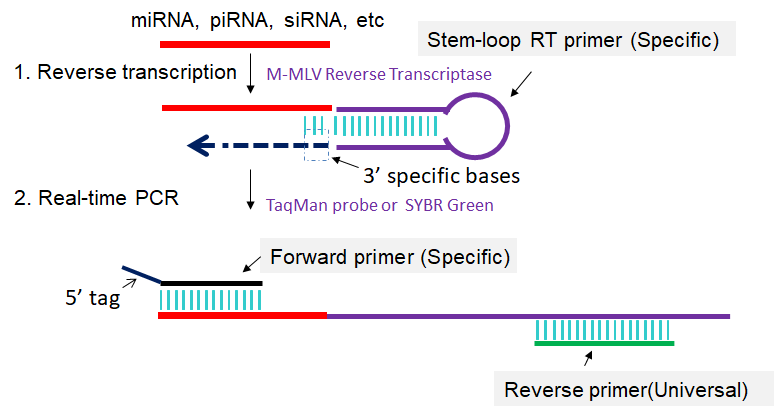
Srnaprimerdb Official Site
Chemicals Used In Pcr

What Are The Role Of Primers In A Polymerase Chain Reaction Quora
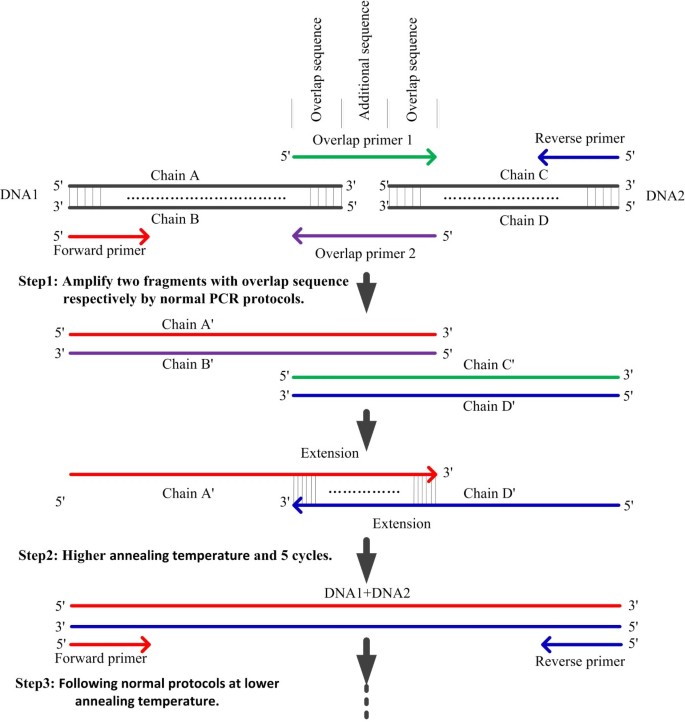
Simultaneous Splicing Of Multiple Dna Fragments In One Pcr Reaction Biological Procedures Online Full Text

Polymerase Chain Reaction Pcr Diamantina Institute University Of Queensland

A Multiplex Rt Pcr For The Detection Of Potato Yellow Vein Virus Tobacco Rattle Virus And Tomato Infectious Chlorosis Virus In Potato With A Plant Internal Amplification Control Wei 09
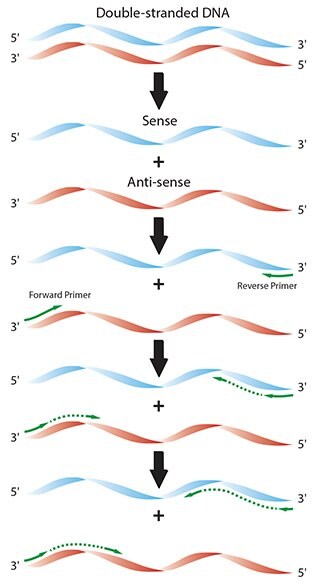
Polymerase Chain Reaction Pcr Process Guide Sigma Aldrich

Kompetitive Allele Specific Pcr Wikipedia
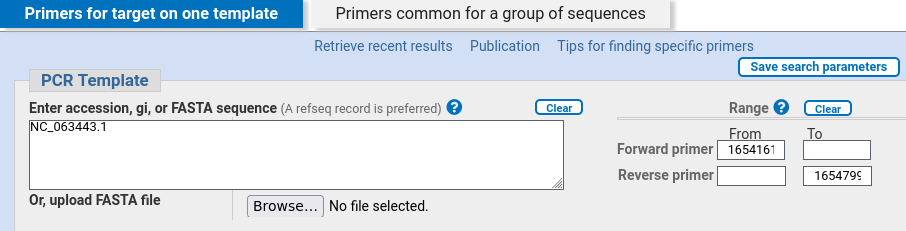
Birch Designing Pcr Primers To Amplify A Gene From Genomic Dna
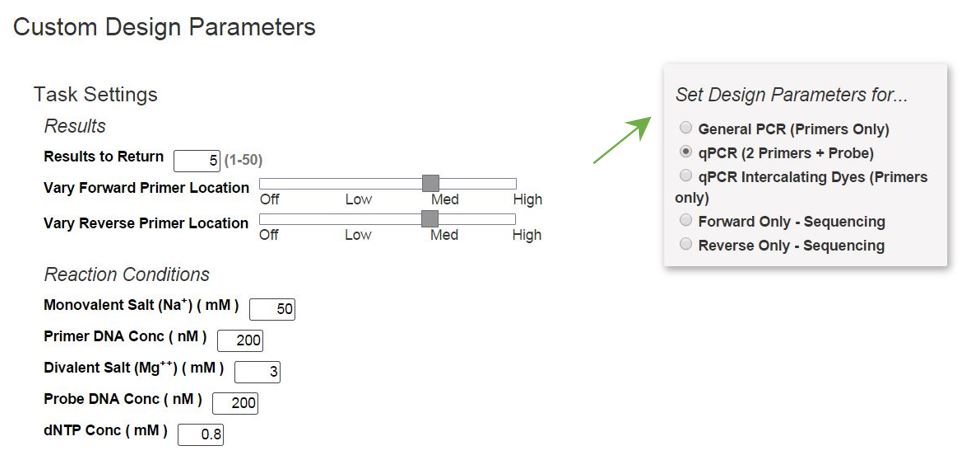
Using Primer Design Tools For Pcr Qpcr Idt

Primerview Forward And Reverse Primer Design From Multi Sequence Datasets Rna Seq Blog

Solved Question 1 The Diagram Below Shows Two Template S Chegg Com
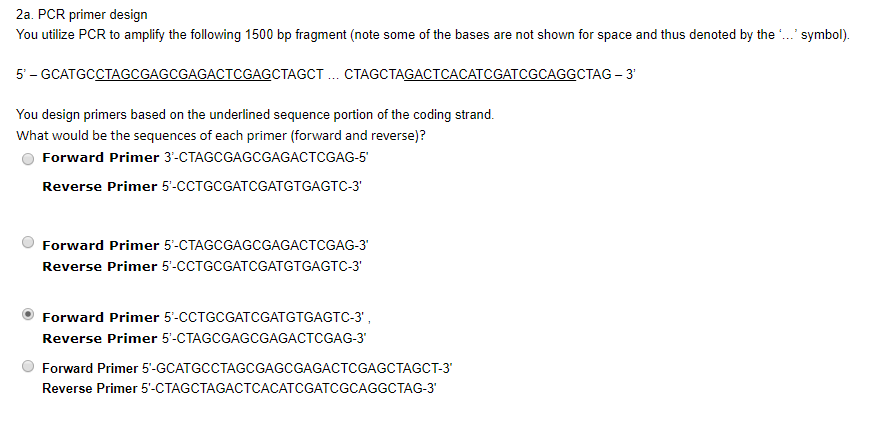
Solved 2a Pcr Primer Design You Utilize Pcr To Amplify T Chegg Com
2

Barcoded Library Preparation Strategy Forward And Reverse Pcr Primers Download Scientific Diagram

Overhang Pcr

Forward And Reverse Primers Used For Rt Pcr Amplification Download Table
Primer Design
Q Tbn 3aand9gcsbd2tmrow Oqz1mdagx7m Jgo9gpxhchw8p Rmy39wululqxpl Usqp Cau

Solved Which Pcr Primer Pair Forward And Reverse Primer Chegg Com
Backpack Pcr A Point Of Collection Diagnostic Platform For The Rapid Detection Of Brugia Parasites In Mosquitoes
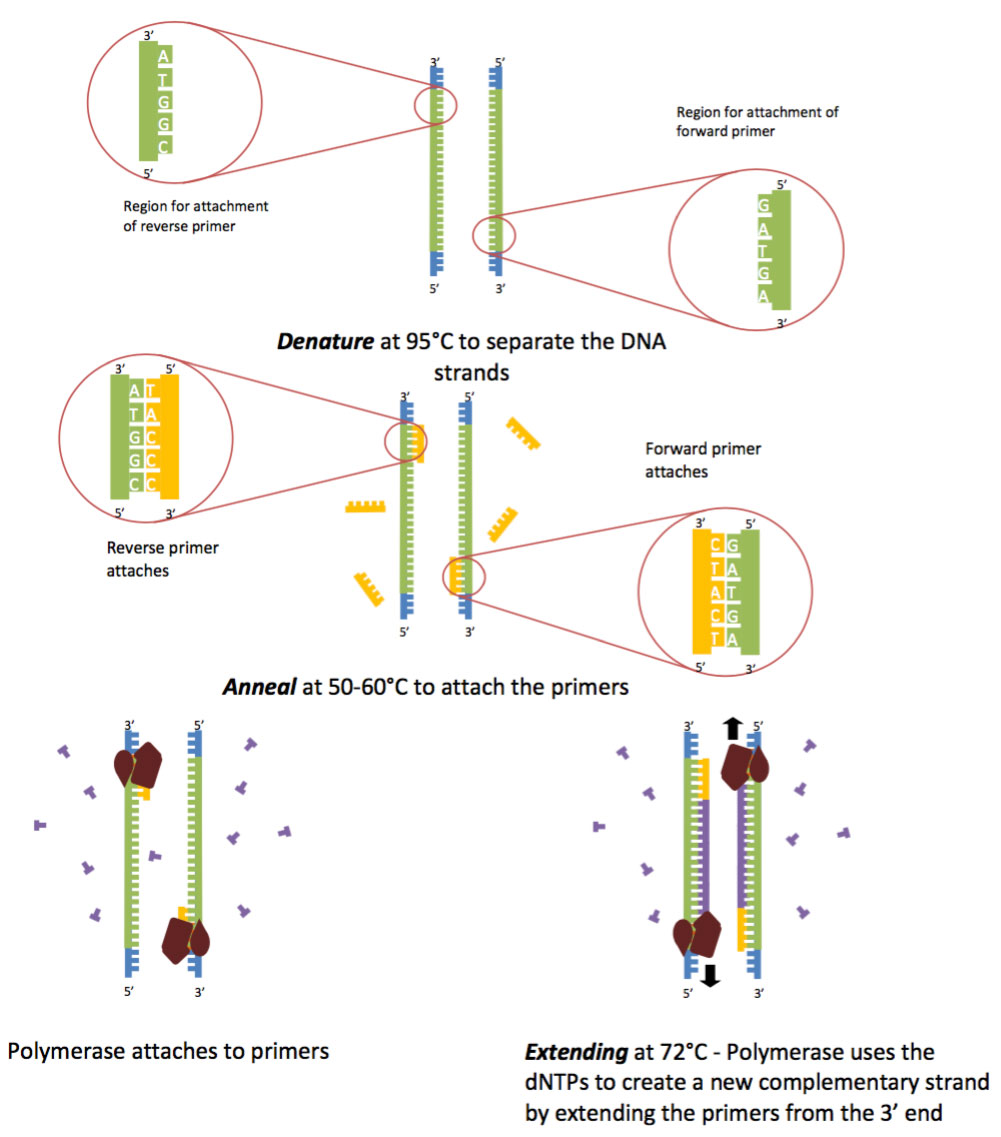
Polymerase Chain Reaction Pcr Diamantina Institute University Of Queensland

Pcr For Sanger Sequencing Thermo Fisher Scientific In
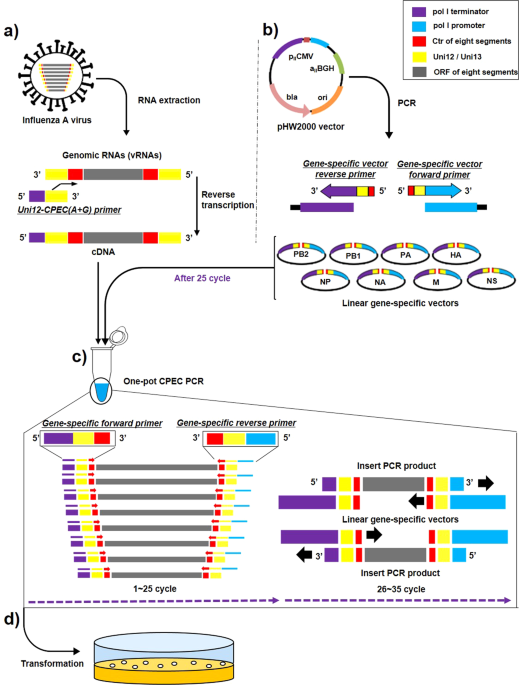
Development Of A Rapid Simple And Efficient One Pot Cloning Method For A Reverse Genetics System Of Broad Subtypes Of Influenza A Virus Scientific Reports

Addgene Plasmid Cloning By Pcr With Protocols

Simera Modelling The Pcr Process To Simulate Realistic Chimera Formation Biorxiv

Janet B Matsen Guide To Gibson Assembly Openwetware
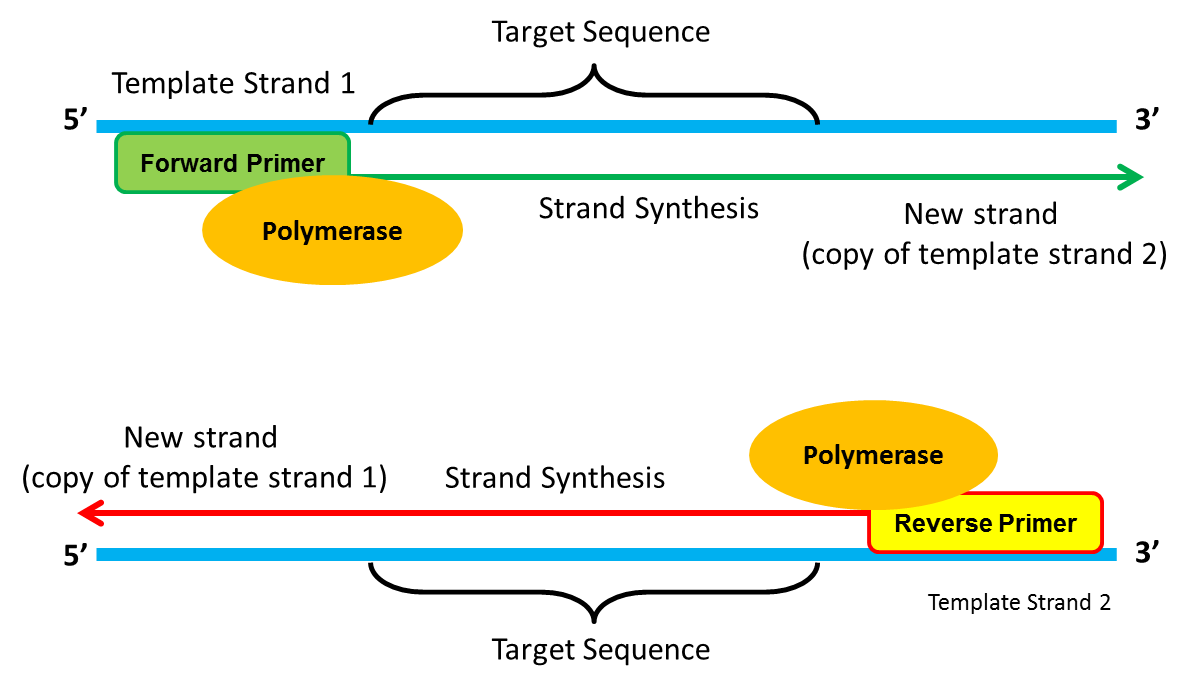
A Cornerstone Of Molecular Biology The Pcr Reaction Antisense Science

Blast Pcr Primers Design Youtube
How Can I Trim And Assemble My Forward And Reverse Sanger Sequence For Each Sample In Batch Using Qiagen Clc Genomics Workbench
2
Q Tbn 3aand9gctvfxchhuftpc Zex Bfuwfrm9 C8 Dg3mp9u1pmqfbzgxvz Go Usqp Cau

Novel Dual Labeled Fluorescence Probe Based Assay To Measure The Telomere Length Biorxiv

Basic Principles Of Rt Qpcr Thermo Fisher Scientific Us

Pcr Methods And Technology For Genetic Analysis
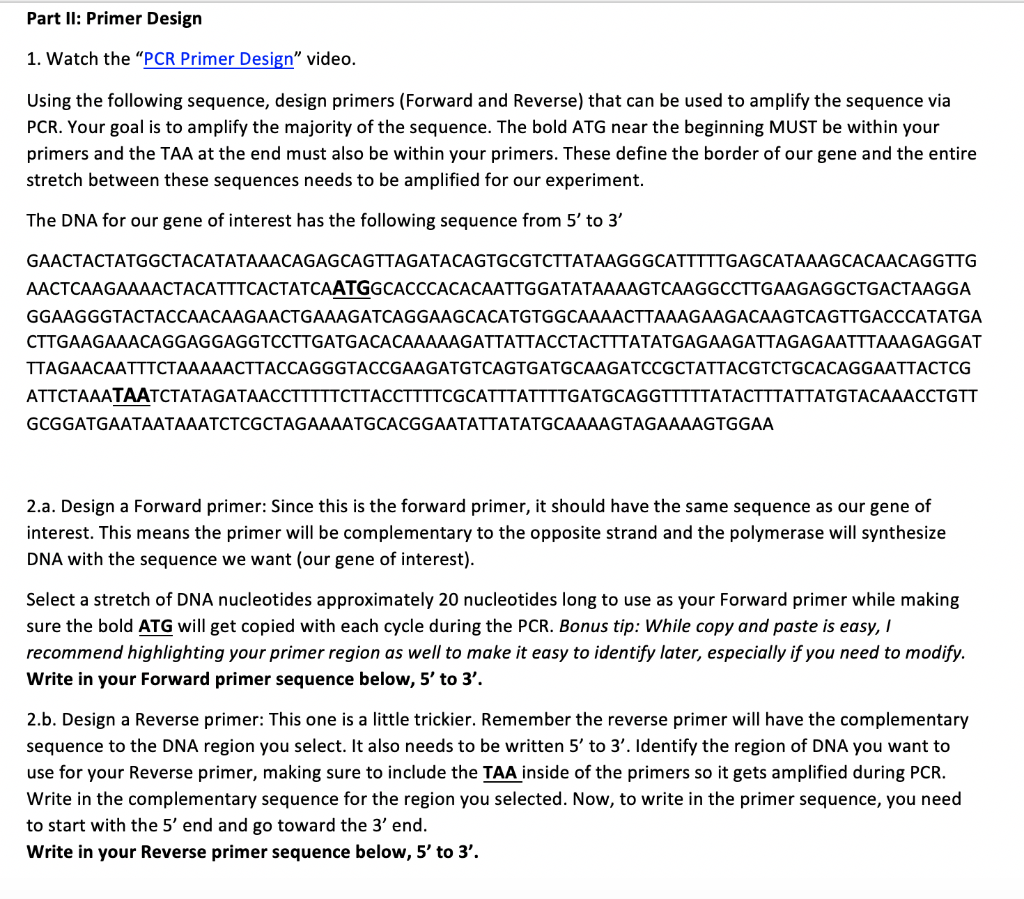
Solved Part Ii Primer Design 1 Watch The Pcr Primer De Chegg Com
Anti Primer Quenching Based Real Time Pcr For Simplex Or Multiplex Dna Quantification And Single Nucleotide Polymorphism Genotyping Nature Protocols

Pcr Primers And Rsai Restriction Enzyme The Forward And Reverse Download Scientific Diagram

Ntmg Gmc Oblab Bioinformatics
What Is The Difference Between Forward And Reverse Primers Pediaa Com
Real Time Pcr Based Assay To Quantify The Relative Amount Of Human And Mouse Tissue Present In Tumor Xenografts Bmc Biotechnology Full Text
In Silico Pcr Primer Design And Gene Amplification Benchling
2

Difference Between Forward And Reverse Primer Compare The Difference Between Similar Terms

A Broadly Applicable Coi Primer Pair And An Efficient Single Tube Amplicon Library Preparation Protocol For Metabarcoding Rennstam Rubbmark 18 Ecology And Evolution Wiley Online Library

Solved You Wish To Design Pcr Primers To Amplify This Reg Chegg Com

Team Sriwijaya Parts 19 Igem Org

Real Time Polymerase Chain Reaction An Overview Sciencedirect Topics
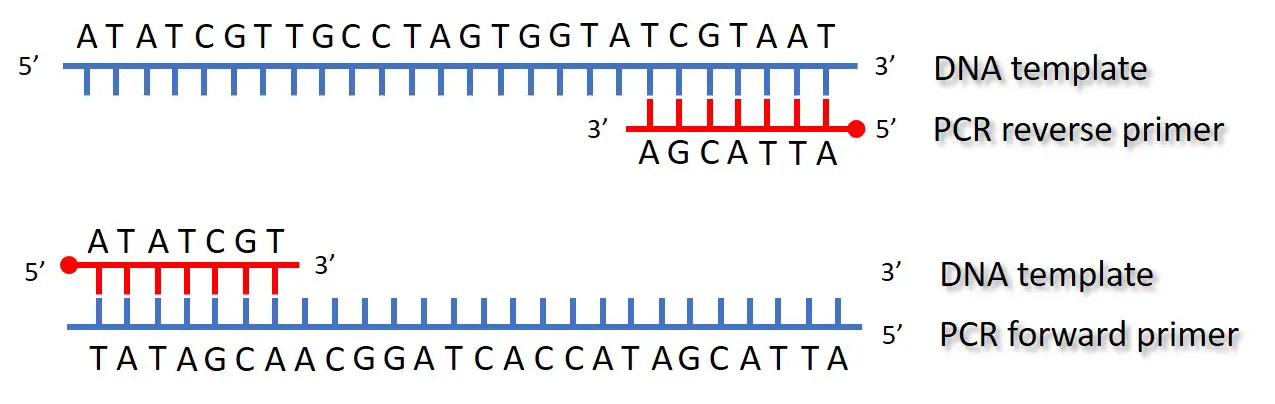
What Is The Polymerase Chain Reaction Pcr

Pcr And Molecular Biology Fundamental Principles
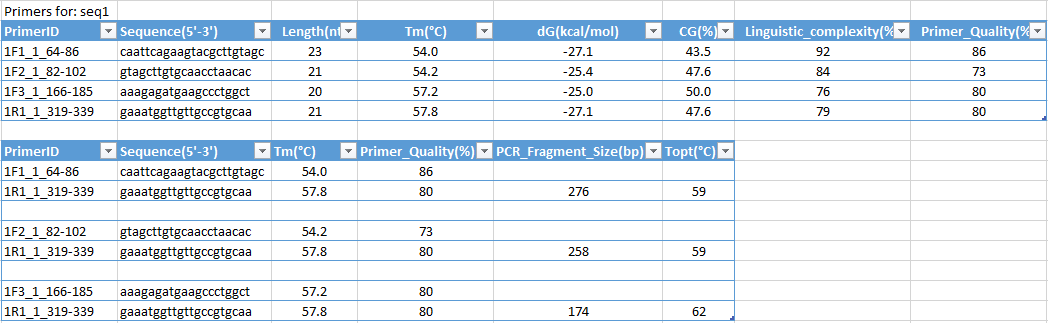
Pcr Primer Design In Silico Pcr And Oligonucleotides

Solved Select The Potentially Good Sets Of Primers For Pc Chegg Com
What Are Forward Primer And Reverse Primer In Pcr Quora

Ncbi Insights Electronic Pcr E Pcr Is Retiring Use Primer Blast Instead
Www Fluidigm Com Binaries Content Documents Fluidigm Scripthub Scripts 16 11 Arguel 16 1 Arguel 16 1 Fluidigm 3aattachments 5b3 5d

Golden Gate Primer Design
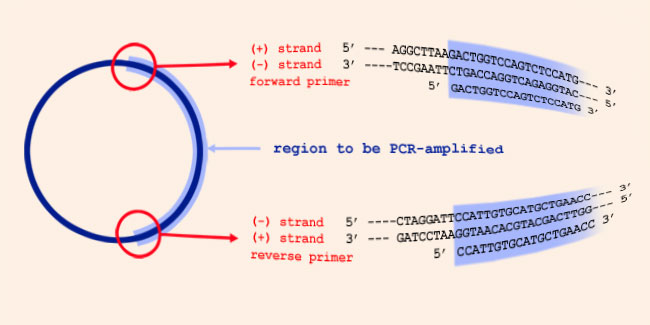
Sequence Notation

In Silico Pcr Unipro Ugene Online User Manual V 1 15 0 Wiki
Q Tbn 3aand9gcrrddijp2sp0zz7qya5zth80pmnqqtbgfztix Bhk0edei36qt3 Usqp Cau

Reverse Transcription Polymerase Chain Reaction An Overview Sciencedirect Topics

Addgene Protocol How To Design Primers

Forward F1 And F2 And Reverse R1 Soma Pcr Primers That Were Used Download Scientific Diagram
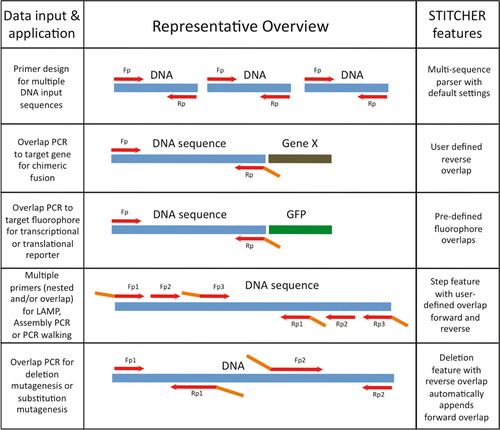
Stitcher A Web Resource For High Throughput Design Of Primers For Overlapping Pcr Applications Biotechniques
Q Tbn 3aand9gcqksw6z5yfg9k6y7nfzbgoh65zaj7edursq Hmswh5zid18pniz Usqp Cau
A Simplified Workflow For Monoclonal Antibody Sequencing

Three Types Of Primer3 Task Arrows Represent Pcr Primers With 3 Ends Download Scientific Diagram

Polymerase Chain Reaction Wikipedia
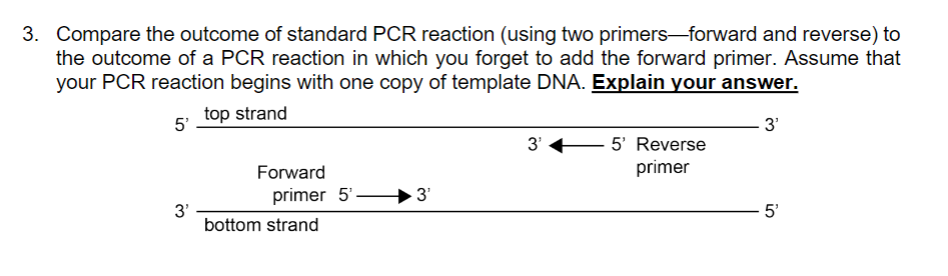
Solved 3 Compare The Outcome Of Standard Pcr Reaction U Chegg Com

Pcr For Sanger Sequencing Thermo Fisher Scientific In

Addgene What Is Polymerase Chain Reaction Pcr

Forward And Reverse Primers Used In The Rt Pcr Reaction To Assess Download Scientific Diagram

The Most Common Questions About Our Indexing Pcr And Sequencing Primer Kit
Journals Plos Org Plosone Article File Type Supplementary Id Info Doi 10 1371 Journal Pone S002
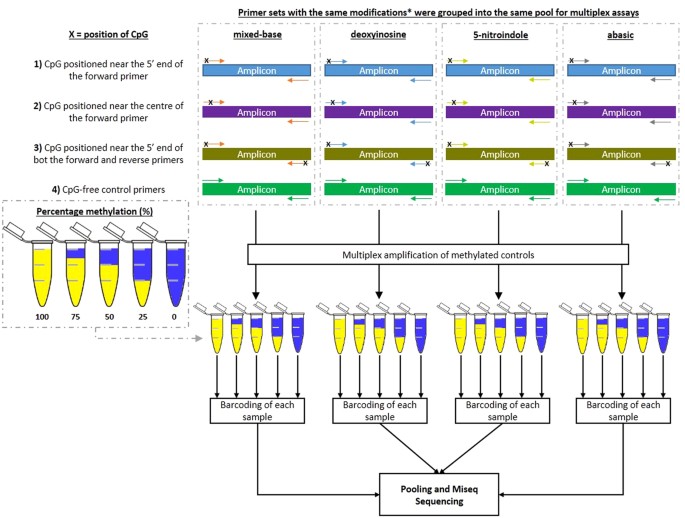
Evaluation Of Different Oligonucleotide Base Substitutions At Cpg Binding Sites In Multiplex Bisulfite Pcr Sequencing Scientific Reports
First Pcr Round Of The Sss Strategy A Two Primers Forward And Download Scientific Diagram

At1 And At2 Angiotensin Receptor Gene Expression In Human Heart Failure Circulation

Primer Design Strategy For The Forward And Reverse Pcr Primers Primers Download Scientific Diagram

Fastpcr Manual

Fastpcr Manual
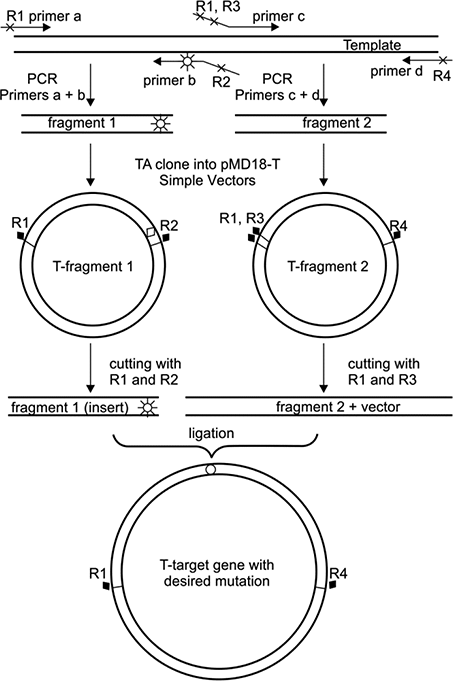
Site Directed Mutagenesis By Polymerase Chain Reaction Intechopen
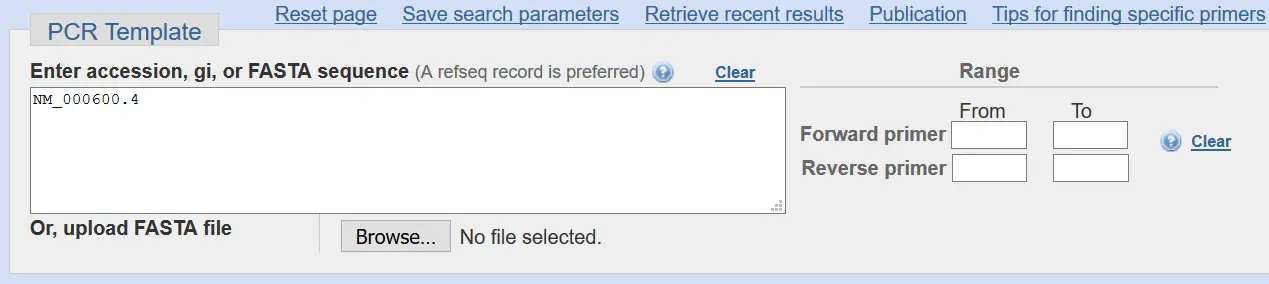
How To Create Real Time Pcr Primers Using Primer Blast

Real Time Pcr Qpcr Primer Design Using Free Online Software Thornton 11 Biochemistry And Molecular Biology Education Wiley Online Library
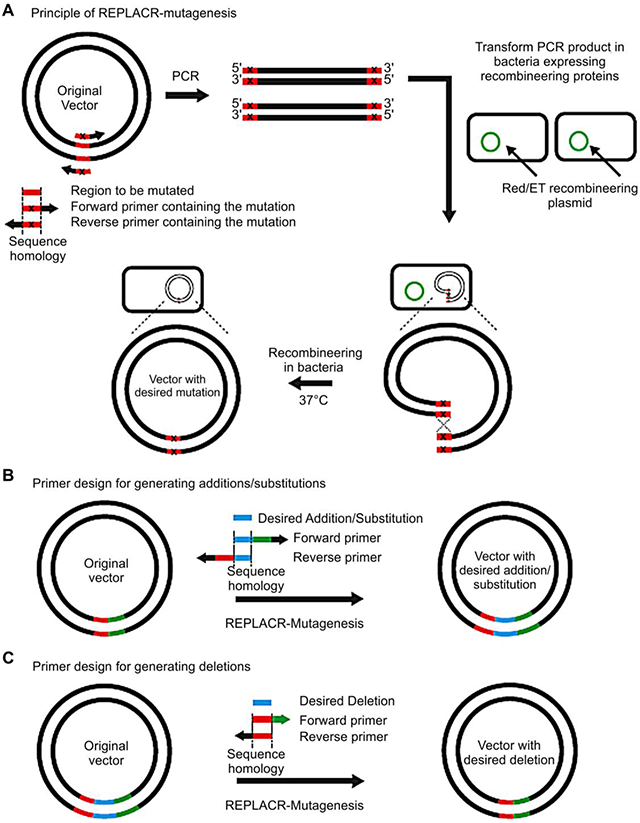
Site Directed Mutagenesis By Polymerase Chain Reaction Intechopen
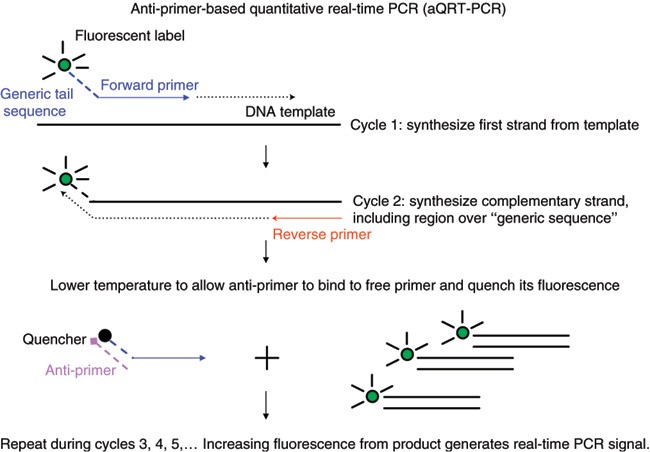
Anti Primer Quenching Based Real Time Pcr For Simplex Or Multiplex Dna Quantification And Single Nucleotide Polymorphism Genotyping Nature Protocols

How To Select Primers For Polymerase Chain Reaction

Novel Dual Labeled Fluorescence Probe Based Assay To Measure The Telomere Length Biorxiv

Forward F1 And F2 And Reverse R1 Soma Pcr Primers That Were Used Download Scientific Diagram

Addgene Plasmid Cloning By Pcr With Protocols
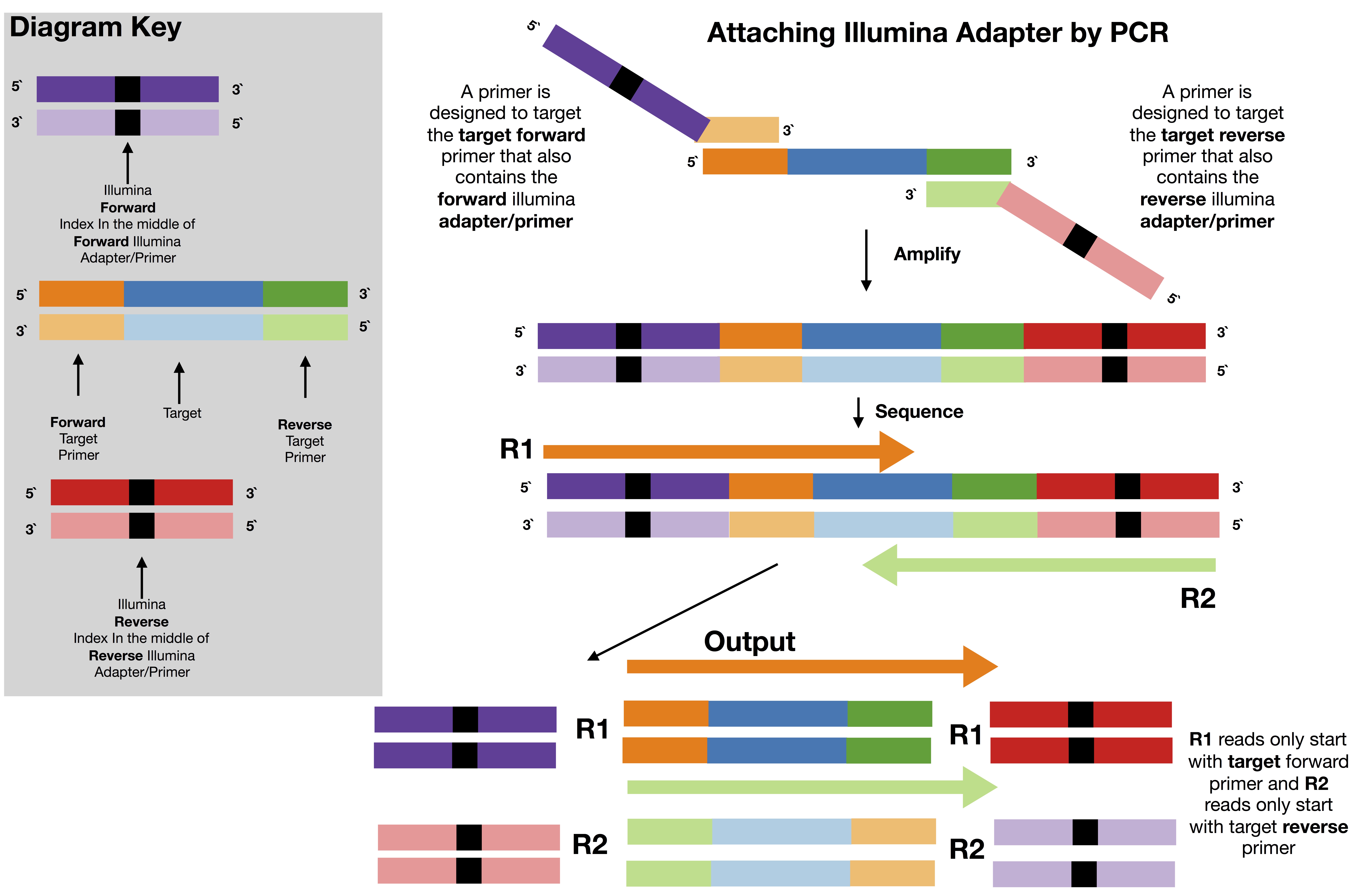
Illumina Paired End Information
Pcr Primers F Forward Primer R Reverse Primer And Size Of Amplicon Download Table

Polymerase Chain Reaction Pcr Diamantina Institute University Of Queensland

Forward And Reverse Primer Sequences Used For Qrt Pcr Download Table

Forward And Reverse Primers Sequences Used In Rt Pcr Download Table
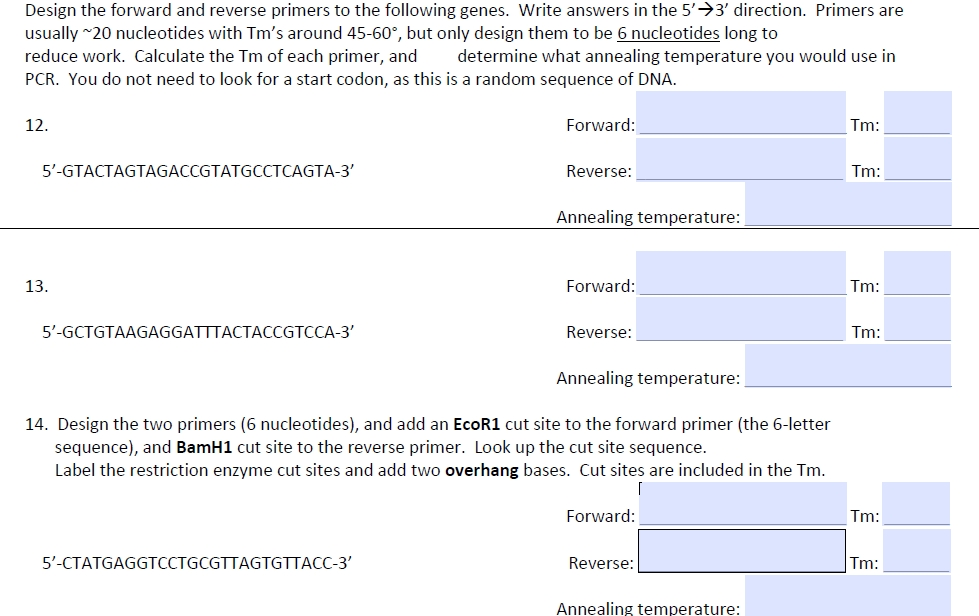
Solved Design The Forward And Reverse Primers To The Foll Chegg Com



