Protein Assay Graph
Hydrolysis is probably the most accuratemethod of determining protein concentration followed by amino acid analysis.
Protein assay graph. Bradford Protein Assay - based on the binding of prot ein molecules to. The bacterial samples of 30 and 60 minutes from lab 6, and 3. Stock of Bradford reagent.
Depending on the sample type and purpose for performing an assay, protein-to-protein variation is an important consideration in selecting a protein assay method and in selecting an appropriate assay standard (e.g., BSA vs. 2 Click twice on the default sheet name in the Prism Navigator tree to rename the sheet. EDTA, Tris, carbohydrates, reducing agents (e.g., DTT, 2-mercaptoethanol), and potassium and.
The Bradford protein assay is a time-tested colorimetric assay. Validation involves a risk assessment step that is followed by a close analysis of the robustness of each assay. I did Bradford assay to get a standard curve in order to determine the protein concentration.
When you graph your protein assay data in Excel, should you include the absorbance of your unknown BSA sample?. 6) BCA Protein Assay:. This protocol allows the preparation of a nine-point serial dilution of BSA standard in order to obtain a standard curve.
The equation for the curve is y = -4x10-5 x 2 + 0.0115x - 0.0027. Cuprous ions are produced by the reduction of cupric ions by proteins in alkaline solutions. The subcellular fractionation of the cauliflower, 2.
But, i don't understand why it was said that the value closer to 1 is a better indicator to show that my standard curve is good to determine the protein concentration. When the Bradford reagent (acidified Coomassie Brilliant Blue G-250) binds to proteins, the dye undergoes a color change in the visible spectrum, with the absorbance maximum moving from 470 to 595 nm. To assay protein samples of unknown concentration.
The Coomassie brilliant blue protein assay, commonly known as the Bradford assay 1, is widely used because of its rapid and convenient protocol as well as its relative sensitivity. Protein concentration is the independent variable (x) and A595 is the dependent variable (y). Explain why or why not.
Add 1 ml of 1 M NaOH with 0.1 ml of diluted sample plus 5 ml of reagent, protein assay 230 Pierce, stirred with a vortex mixer. The Lowry protein assay method for protein concentration determination is one of themost venerable and widely-used protein assays. Standard Curve The Bradford assay is calibrated by using a pure protein of known concentration, called the "standard protein".
View the graph that is produced from the data by clicking Protein Standard Curve under Graphs in the navigator bar. Briefly describe the principles behind the protein assay and their weakness and strengths. The Bradford protein assay is a dye-binding assay based on the differential color change of a dye in response to various concentrations of protein.
Read the absorbance with a spectrophotometer at 595 nm. Calibration curve for BSA standard is prepared using standard albumin, 50 ml Pierce with concentration of 2 g/l, diluted with 1 M NaOH solution. Bradford assay is a protein quantification protocol developed by Marion Mckinley Bradford in 1976.
Image depicts the screen output from KC3 of a typical standard curve of a Lowry protein assay. Unlike BCA, this is an endpoint assay with a stable result, meaning that you can estimate the amount of protein from one assay by comparing it with a previous standard curve!. The Bradford assay is a colorimetric assay that measures protein concentration.
Revised Fall 10 Biuret Protein Assay. 2.1 List of reagents:. Reagents and equipment needed:.
Explain why or why not. # BE -402C ). Pierce 660nm Protein Assay Kit, sufficient reagents for 300 test tube or 3000 microplate assays.
Example of Assay Data Table Create a tablewith the absorbance results obtained from the assay. Most protein assay methods use BSA or immunoglobulin (IgG) as the standard against which the concentration of protein in the sample is determined (Figure 1). Protein standard curve y 0 2 4 6 8 0.00 0.05 0.10 0.15 0.
A protein assay consists of two main components. Without the protein, the solution is red-brown in its acidic solution and when protein binds, the pKa of the dye shifts causing the dye to become blue. Note that the equation describing the regression curve is provided along with statistics concerning the curve.
For greater accuracy run this step in duplicate. The protein sample, irrespective of its composition, is measured through absorbance spectroscopy at 540 nm in conjunction with a k. The Bradford Protein Assay measures protein concentration in a sample.
Enjoy the videos and music you love, upload original content, and share it all with friends, family, and the world on YouTube. Standard curve prepared using the protein standards (Table 2 and Graph 1). It is a sensitive technique.
The Bradford is the reverse:. Prepared from the protein and assayed alongside the unknown(s) before the concentration of each unknown is determined based on the standard curve. Asked Sep 18, 19.
It is a quick and accurate spectroscopic analytical procedure used to measure the concentration of protein in a solution. Biuret Protein Assay Teachers Hand book (Cat. Like other protein assays, the result of the Bradford assay differs for different proteins.
To know determine the actual. The Coomassie brilliant blue protein assay, commonly known as the Bradford assay 1, is widely used because of its rapid and convenient protocol as well as its relative sensitivity. The absorbance is measured using a spectrophotometer, at the maximum absorbance frequency (A max) of the blue dye (which is 595 nm).
Absorbance was measured at 750 nm using visible spectrophotometry, using 0.4 mg/mL BSA as the protein standard. Briefly describe the principles behind the protein assay and their weakness and strengths. A seven-point standard curve for the estimation of target protein amount by the Lowry method.
This assay works by measuring the color change achieved with the basic amino acids combined with Coomassie dye, which, under. By measuring absorbance values corresponding to a set of protein standards we can plot a standard curve of absorbance versus amount of protein. The absorbance readings obtained from these solutions are used to construct a graph of absorbance as a function of protein concentration.
Linearity of the assay. Through this curve you can determine the unknown protein concentration by use of Beer’s Law. When performing ELISAs, three different data outputs can be generated:.
A standard protein curve is made to show the relationship of the concentration of the protein to that of the absorbency at 595nm. The standard curve and the unknowns. + bx + c, where solving for x determines the protein concentration of the sample.
Pierce 660nm Protein Assay Reagent, 450mL. About twice as sensitive to BSA as to IgG. The Standard assay is used to measure protein concentrations within a working range of 10 to 00 μg/mL in a 96-well plate.
-we are going to do the protein assay for three samples:. The BCA Protein Assay has been advertised as an alternative to the Lowry assay. µg protein added 0 2.5 5 7.5 10 12.5 15 17.5 25 30 35 2mg/mL BSA (μL) 0 1.25 2.5 3.75 5 6.25 7.5 8.75 10 12.5 15 17.5 H.
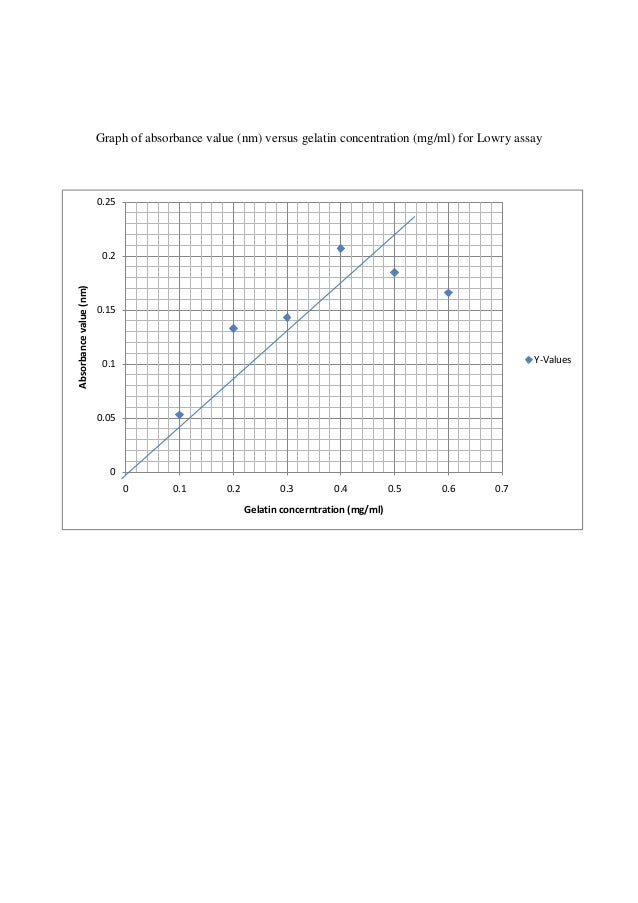
Unfortunately, this assay isn’t compatible with lots of common chemicals:. The Bradford is recommended for general use, especially for determining protein content of cell fractions and assesing protein concentrations for gel electrophoresis. Graph of absorbance value (nm) versus gelatin concentration (mg/ml) for Biuret assay 0.25 0.
# BE -402B ). Slightly exceed the standard concentration of gelatin. Page 6 of 8.
Unfortunately, there is a large degree of curvature over a broad range of protein concentrations (Fig. In the Bradford assay, we used the dye Coomassie G-250 which binds to proteins mostly at arginine but also at tryptophan, tyrosine, histidine and phenylalanine residues (Olson, 07). 0.04mg 1mg BSA protein used the linear part of the curve.
A protein assay consists of two main components:. Protein + standard Linear (Standard alone) Linear (Protein + standard) Assay 1) Prepare Standards as indicated below in glass tubes (16 x 100 mm). The data below should not be used as a replacement of a standard curve.
Linearization of the Bradford Protein Calibration Graph:. Absorbance of protein (raw fish) = 0.280 A y = 0.1276 ln (x) + 0.0346 ln x = 0.280 – 0.0346 0.1276 x = 6.84 mg/ml (protein concentration) We have plotted the graph for the standard gelatin and raw fish protein. As each protein requires a tailored quantification method this will to some extent.
If precise quantitation of an unknown protein is required, it is advisable to select a protein. Coomassie Protein Assay Teacher ï book (Cat. To analyze data from standard curve and unknowns.
Linearization of the Bradford Protein Calibration Graph:. Bradford and Lowry protein assay reagents results in a change in absorbance when protein is present. The enzyme-linked immunosorbent assay (ELISA) is a sensitive technique for determining the concentration of a protein in a liquid sample.
This graph is called the standard curve for assay, and can be used to convert the absorbance readings for the experimental samples (eggs from duck, chicken 1 (ayam biasa), chicken 2 (ayam kampung), omega 3. How does this absorbance relate to the actual protein concentration?. However, if great accuracy is required, prepare the standard curve from a pure sample of the target protein.
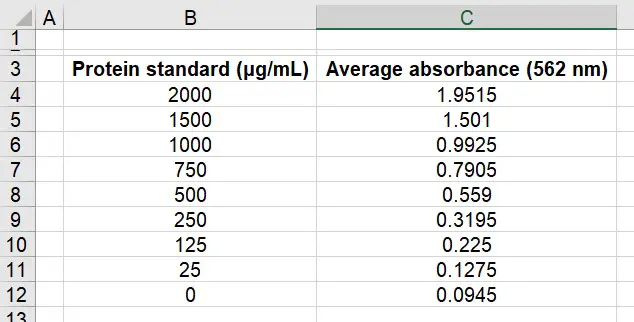
Concentration curve from 0 to µg/well of BSA with polynomial regression analysis. The key component in this assay is bicinchoninic acid (BCA) which reacts with cuprous ions to generate an intense purple color at 562 nm. Because the color response with coomassie is non-linear with increasing protein concentration, a standard curve must be completed with each assay.
How the Bradford Protein Assay Works. Reference to absorbances obtained for a series of standard protein dilutions, which are assayed alongside the unknown samples. Bicinchoninic acid assay (Protein.
To pipet accurately. The Bradford protein assay was developed by Marion M. It is fairly accurate and samples that are out of range can be retested within minutes.
Bovine Serum Albumin (BSA) Set, 7 × 3.5mL, contains. The reagent Coomassie Brilliant Blue turns blue when it binds to arginine and aromatic amino acids present in proteins, thus increasing the absorbance of the sample. The absorbance of the BSA protein standards (Tubes 1–5) in each assay.
Stock solution of Bovine Serum Albumin (BSA):. It gives an easy way to estimate the protein concentration of a solution using a standard curve generated by the use of known concentrations of a protein. If the absorbances of the final solutions (green) are identical, then the concentration of the unknown sample is determined to be 1 mg/mL.
Pre-diluted Protein Assay Standards:. 0129.7 230 Coomassie (Bradford) Protein Assay Kit. The Bradford assay relies on the binding of the dye Coomassie Brilliant Blue G250 to protein, in which the dye is proportional to the protein concentration.
Use graph paper or a computer program like Excel to create a scatter plot of the Bradford protein assay data for the BSA parallel dilutions 1-7. While doing a protein assay throughout this semester. -Graph known amount of protein vs.
5000 microplate assays. The standard curve and the unknowns. The dye reagents are commonly purchased from Bio-Rad (Richmond, CA).
(absorbance) for the standards → we are going to create a standard curve and determine the slope of the line. The linear graph acquired from the assay (absorbance versus protein concentration in μg/mL) can be easily extrapolated to determine the concentration of proteins by using the slope of the line. Protein assay methods based on similar chemistry have similar protein-to-protein variation.
Biuret Protein Assay - based on binding of copper ions to peptide bonds under. Bradford and Lowry protein assay reagents results in a. • Qualitative.An ELISA can be used to determine whether the protein of interest is found within a particular sample by comparing the sample to a blank well or a sample that does not.
In assays using 5 ml color reagent prepared in the lab, the sensitive range is closer to 5 to 100 µg protein. If standard (top row) and unknown (bottom row) samples are dispensed and mixed with the same amount of assay reagent, then they are directly comparable. To prepare a standard curve.
This is if the standard curve is linear, however, with BSA standard curve (x axis is time starting from 0) it is not linear and is not that simple. In some research applications, this assay is recommended as a replacement for other protein assays, especially the widely used. When you graph your protein assay data in Excel, should you include the absorbance of your unknown BSA sample?.
The Biuret Assay, also known as the Piotrowski Test, is a biochemical assay that allows one to accurately quantify protein concentration within the range of 5-150 mg/mL. You are aware that A 280 is about twice as sensitive to IgG as to BSA. When the dye is allowed to interact with a substance that contains protein, the arginine groups of the protein bind to Coomassie.
Create the BSA protein standard curve from your Bradford protein assay data:. From the graph, the protein concentration in raw fish obtained was 6.84 mg/ml. The Bio-Rad protein assay is a simple colorimetric assay for measuring total protein concentration and is based on the Bradford dye-binding method (Bradford 1976).
Absorbance and amount of protein are continuous variables, so we should add a trend line that relates absorbance to amount over the entire usable range of the assay. Diagram of protein assay steps. The Bradford assay is very fast and uses about the same amount of protein as the Lowry assay.
Unfortunately, there is a large degree of curvature over a broad range of protein concentrations (Fig. 0.005mg t 0.050mg BSA protein used, the linear part of the curve. While doing a protein assay throughout this semester.
Pierce™ BCA Protein Assay Kit. To learn the use of the spectrophotometer.

Pdf Linearization Of The Bradford Protein Assay Semantic Scholar

Standard Curve An Overview Sciencedirect Topics
Bradford Protein Assay Wikipedia
Protein Assay Graph のギャラリー
Http Webservices Itcs Umich Edu Drupal Allenlab Sites Webservices Itcs Umich Edu Drupal Allenlab Files Allen lab bca assay protocol Pdf
How To Create A Linear Standard Curve In Excel

Standard Curve Wikipedia
2

Protein Concentration Using Excel 16 Youtube

Assays For Determination Of Protein Concentration Olson 07 Current Protocols In Protein Science Wiley Online Library

a Assay For Protein Determination Nfsc 450
Solved In Biochemistry We Use A Bradford Assay To Calc Chegg Com

Standard Curve For The Determination Of Protein Concentration By Biuret Download Scientific Diagram

Figure 8 From Linearization Of The Bradford Protein Assay Semantic Scholar

Linear Range Of Detection And What It Means For Your Quantifications Azure Biosystems

Protein Analysis Using Microplate Based Quantification Methods June 28 18
Assets Thermofisher Com Tfs Assets Lsg Manuals Man Mod Lowry Protein Asy Ug Pdf

Log Versus Log Scale Plot Of A Standard Curve For Results From 10 Download Scientific Diagram
Use The Spectrovis Plus To Study Proteins

Calibration Graph For Protein Using The Bradford Assay For Smartphone Download Scientific Diagram
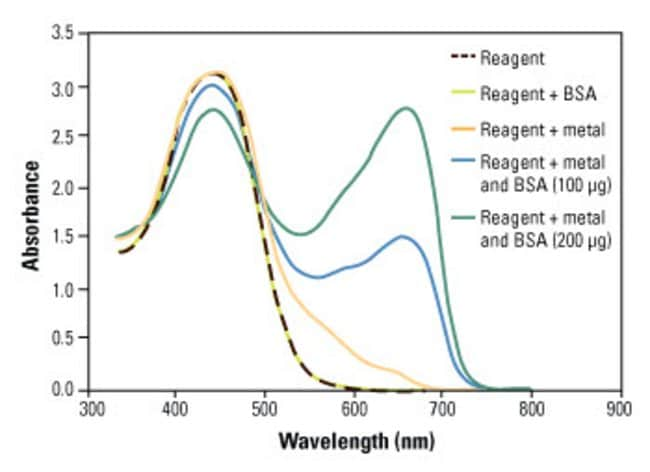
Pierce 660nm Protein Assay Reagent
Q Tbn 3aand9gcq8qafndghkgwuy2syn7t3eh3kbhvyjuxtqgeneq4i 2kezwka6 Usqp Cau
Protein Assay

Linearization Of The Bradford Protein Assay Protocol

Assays For Determination Of Protein Concentration Olson 07 Current Protocols In Protein Science Wiley Online Library

Responses Of Different Proteins In The Bradford Assay

Protein Analysis Using Microplate Based Quantification Methods June 28 18
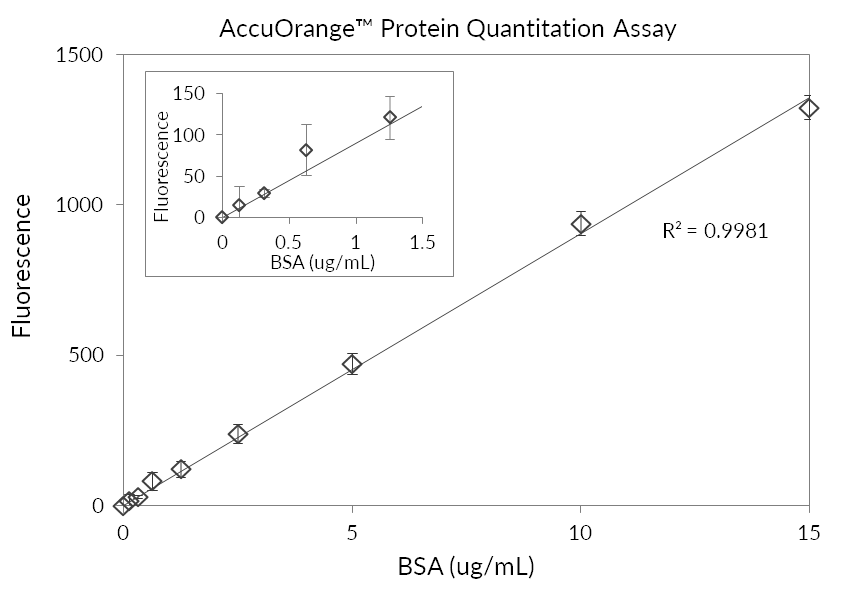
Accuorange Protein Quantitation Kit Biotium

Linearization Of The Bradford Protein Assay Protocol
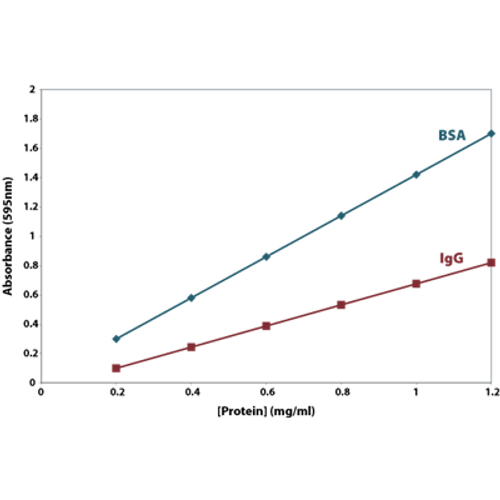
Cb Protein Assay Protein Quantification Assay Protein Research
Q Tbn 3aand9gcqqpwmbrq0qz8qo1g7 7 Umleizjlvestldc3nc1desq2lnbf0a Usqp Cau
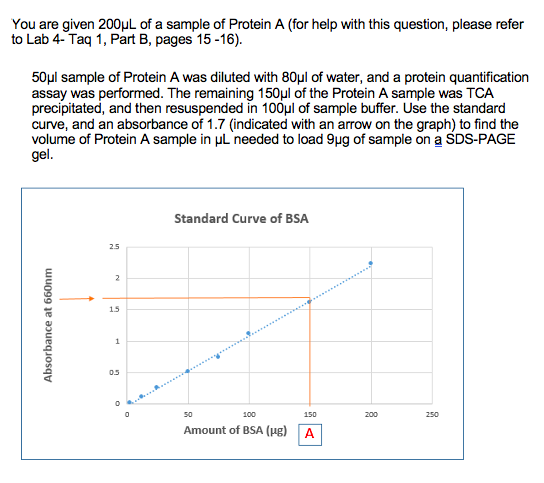
Solved You Are Given 0µl Of A Sample Of Protein A For Chegg Com
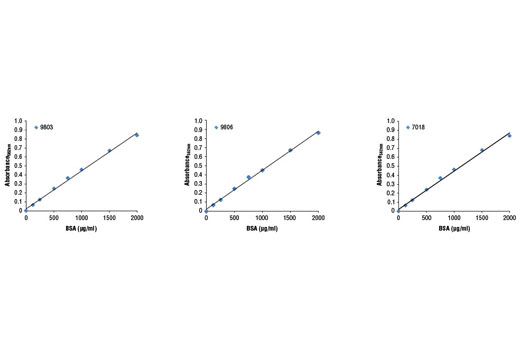
a Protein Assay Kit

Protein Assay Lab Report Jane S Ap Bio Webpage

Bradford Protein Assay Calculation Of An Unknown Standard Protein Bradford Standard

A Analysis Graph Of Total Protein Quantities Of All Groups Detected Download Scientific Diagram

Nick S Teaching Blog Why Can T I Extrapolate The Bradford Assay Graph If The Beer Lambert Law Applies
2

Linearization Of The Bradford Protein Assay Protocol
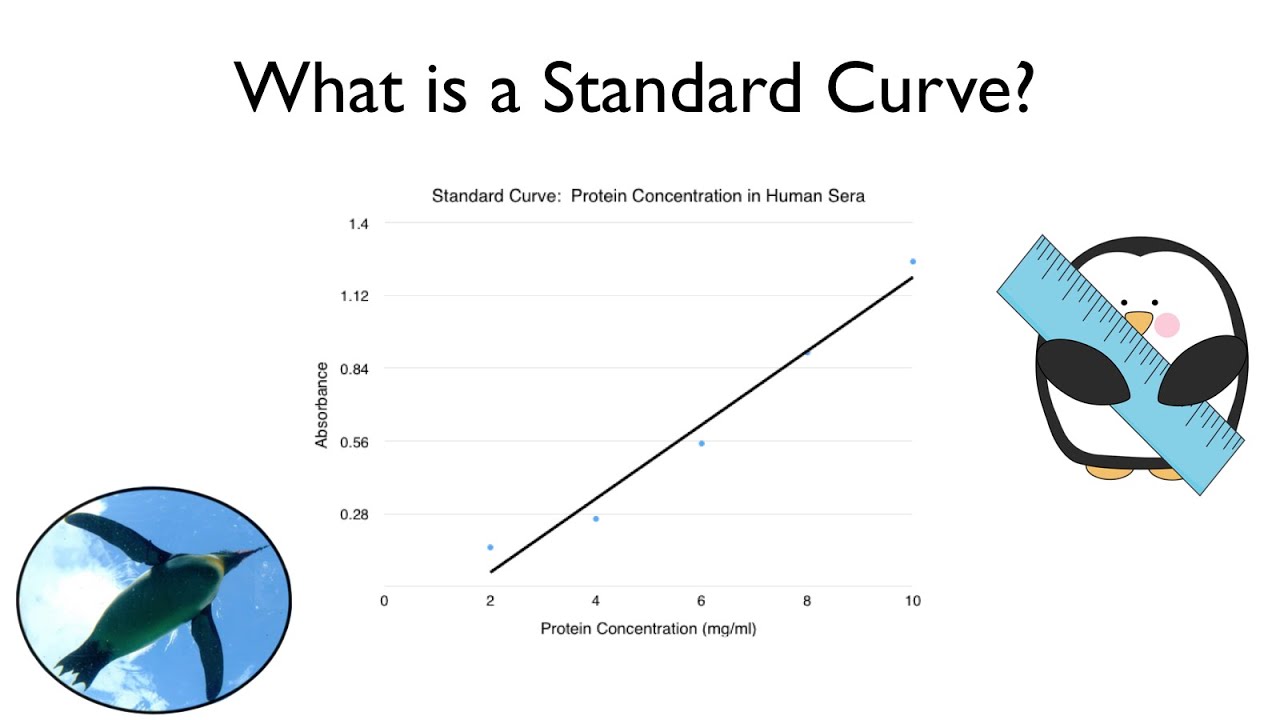
What Is A Standard Curve Youtube

Protein Assay Lab Vannimah

Sensitive Protein Assay Overcomes All Interfering Agents No Protein To Protein Variation

Linearization Of The Bradford Protein Assay Protocol

a Protein Assay Kit Ab Abcam

Assays For Determination Of Protein Concentration Olson 07 Current Protocols In Protein Science Wiley Online Library
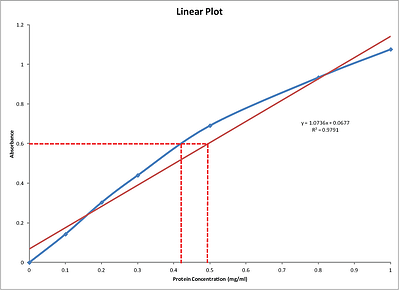
Bradford Protein Assay Calculation Of An Unknown Standard

Biochemistry Practical 2 Protein Experiment
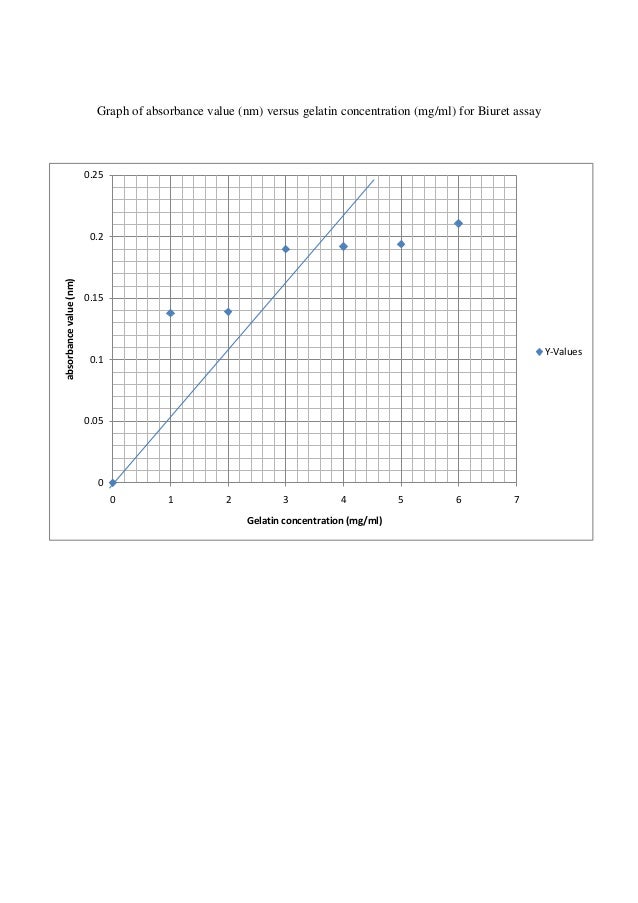
Lowry Vs Biuret Final 1
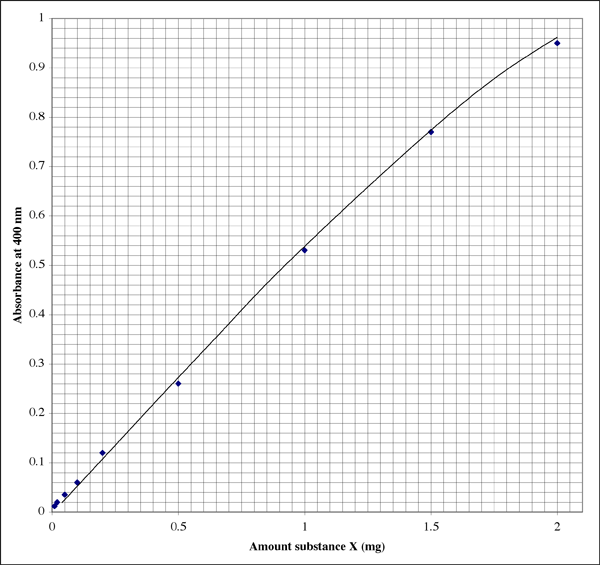
Setting Up A Colorimetric Assay

Prism 3 Calculating Unknown Concentrations Using A Standard Curve Faq 1753 Graphpad

Bradford Assay Calculations Youtube

Pierce a Protein Assay Protocol
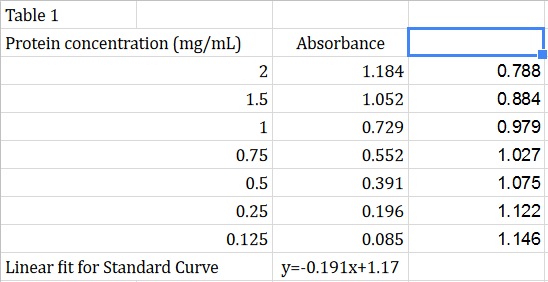
Solved Bradford Assay Lab Compare Your Protein Concentrat Chegg Com

Standard Curve An Overview Sciencedirect Topics
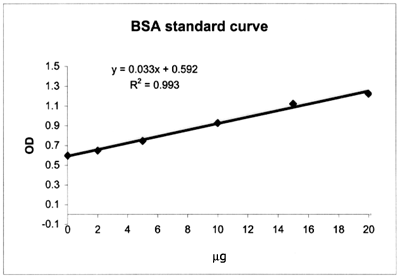
Molecular Endocrinology Laboratory Spectrophotometry

Assays For Determination Of Protein Concentration Olson 07 Current Protocols In Protein Science Wiley Online Library

G Biosciences Protein Assay Cb 500 Assays Protein Assay Cb 500 Assays Fisher Scientific
Q Tbn 3aand9gctwaf5pjnd Ytgtthkdm6yio6vgnvstjzputfjtg4zu9p7nlo15 Usqp Cau
2

Bradford Protein Assay Wikipedia
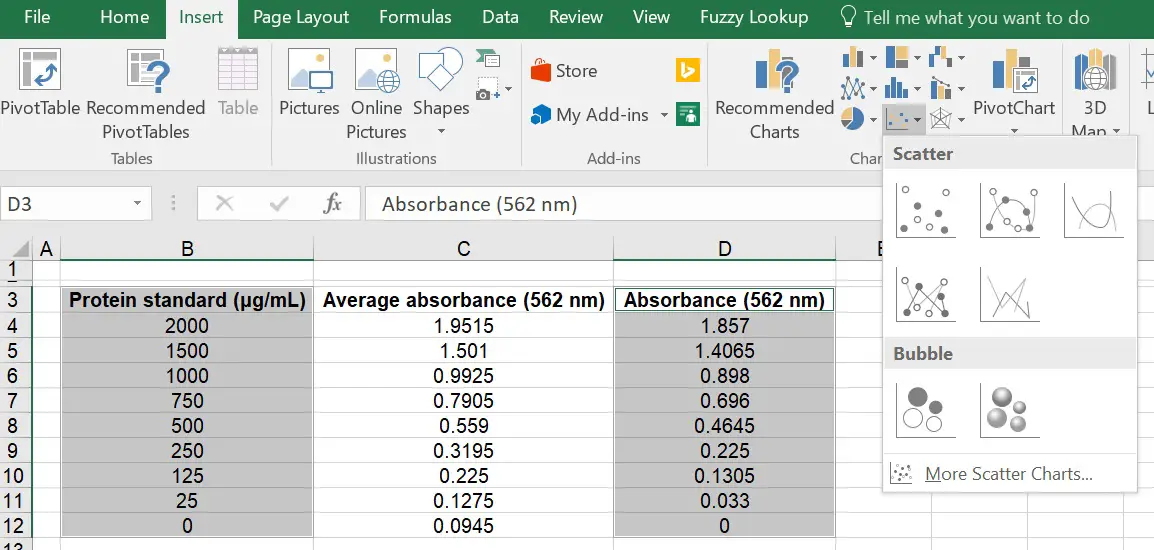
How To Create A Linear Standard Curve In Excel

Graph Showing Linear Plot Of Bsa Standards Used In Protein Download Scientific Diagram

Bradford Protein Assay Wikiwand
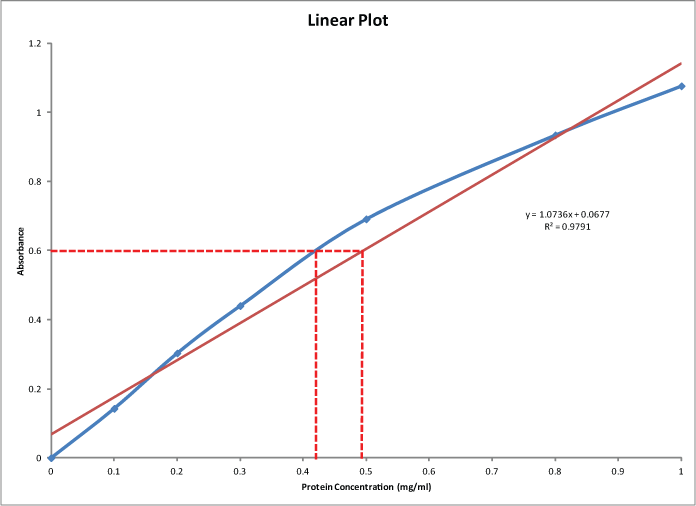
Lowry Vs Biuret Final 1

Amplite Fluorimetric Fluorescamine Protein Quantitation Kit Blue Fluorescence t Bioquest

Standard Curve An Overview Sciencedirect Topics
Bradford Protein Assay Calculation Of An Unknown Standard

Protein Measurement Find A Suitable Method Bmg Labtech
2
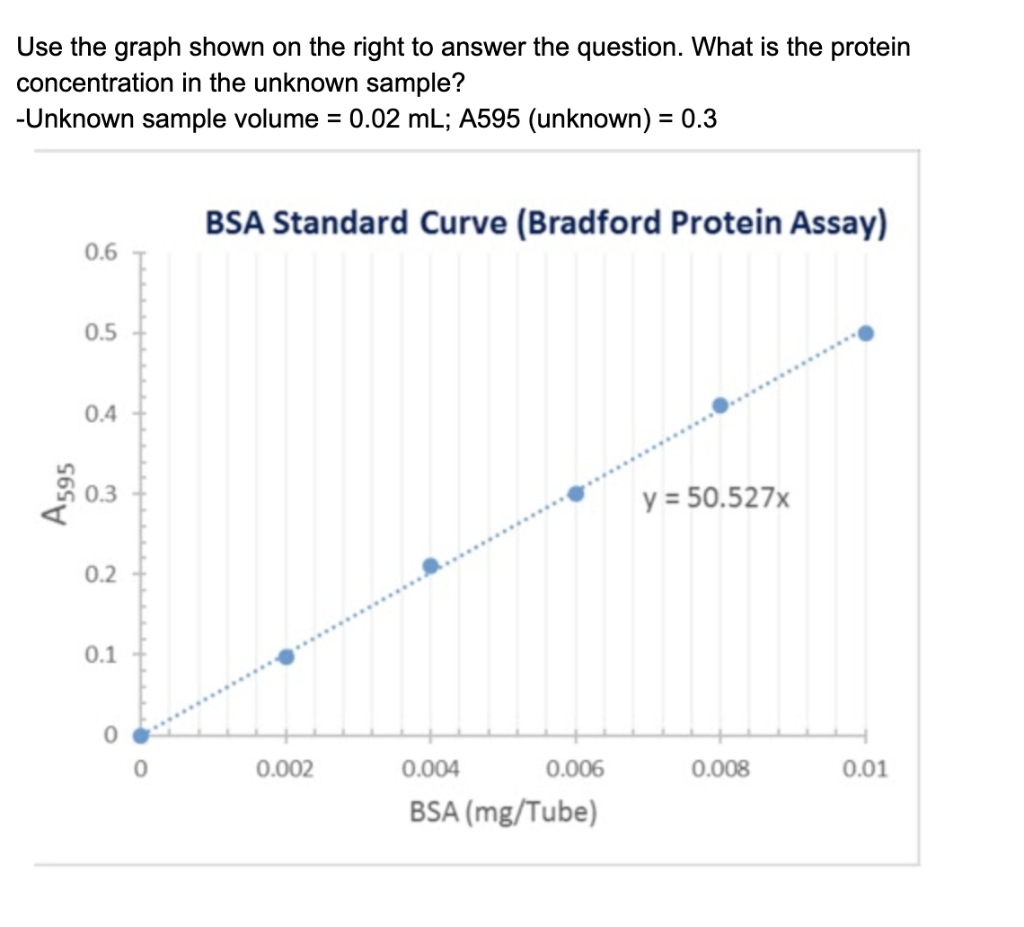
Solved Use The Graph Shown On The Right To Answer The Que Chegg Com
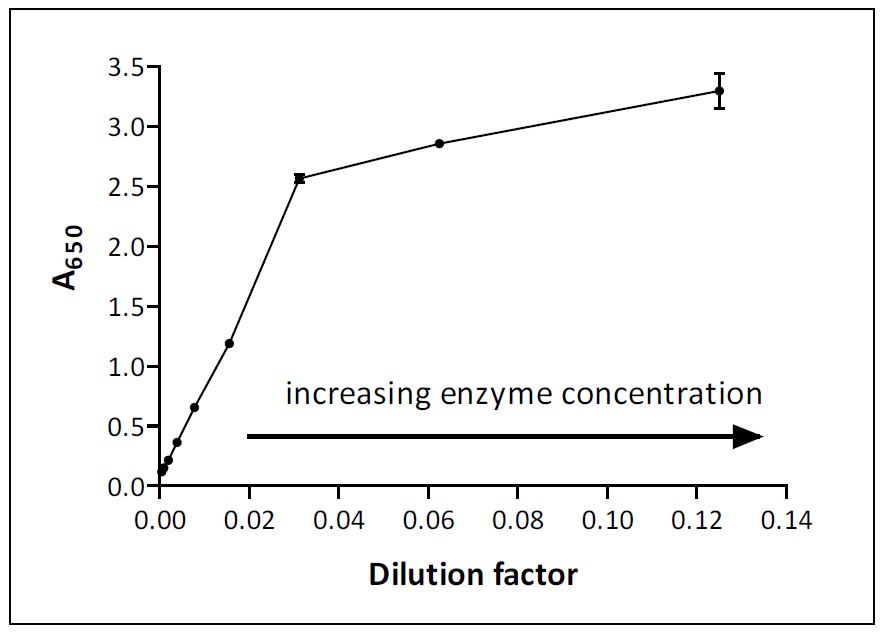
Guide To Enzyme Unit Definitions And Assay Design Biomol Blog Resources Biomol Gmbh Life Science Shop

Figure 4 From Linearization Of The Bradford Protein Assay Increases Its Sensitivity Theoretical And Experimental Studies Semantic Scholar

The Use Of Multiplex Platforms For Absolute And Relative Protein Quantification Of Clinical Material Sciencedirect

Calibration Graph For Protein Using The Bradford Assay For Smartphone Download Scientific Diagram

Quantification Of Soluble Or Insoluble Fractions Of Leishmania Parasite Proteins In Microvolume Applications A Simplification To Standard Lowry Assay

B Bsa Calibration Curve This Standard Curve Of Protein Concentration Download Scientific Diagram

G Biosciences Cb X Protein Assay 500 Assay Cb X Protein Assay 500 Assay Fisher Scientific

Bradford Protein Assay Principle Protocol Calculations Video Lesson Transcript Study Com

a Protein Assay Kit Ab Abcam

Cb X Protein Assay With Bsa Standard From G Biosciences Biocompare Com

Protein Assay Writing Assignment Graph Guidelines Youtube
Lab 6 Protein Quantification Biology1

Bradford Assay Kit Ab Abcam

Figure 5 From Linearization Of The Bradford Protein Assay Increases Its Sensitivity Theoretical And Experimental Studies Semantic Scholar

R For Biochemists Let S Analyse A Protein Assay
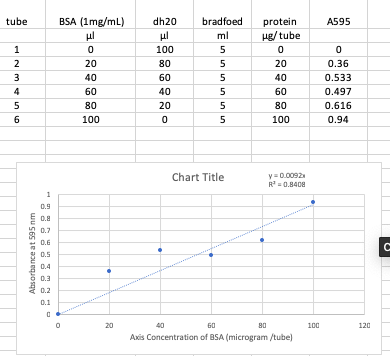
Solved Calculate The Protein Ug Tube From The Graph Belo Chegg Com
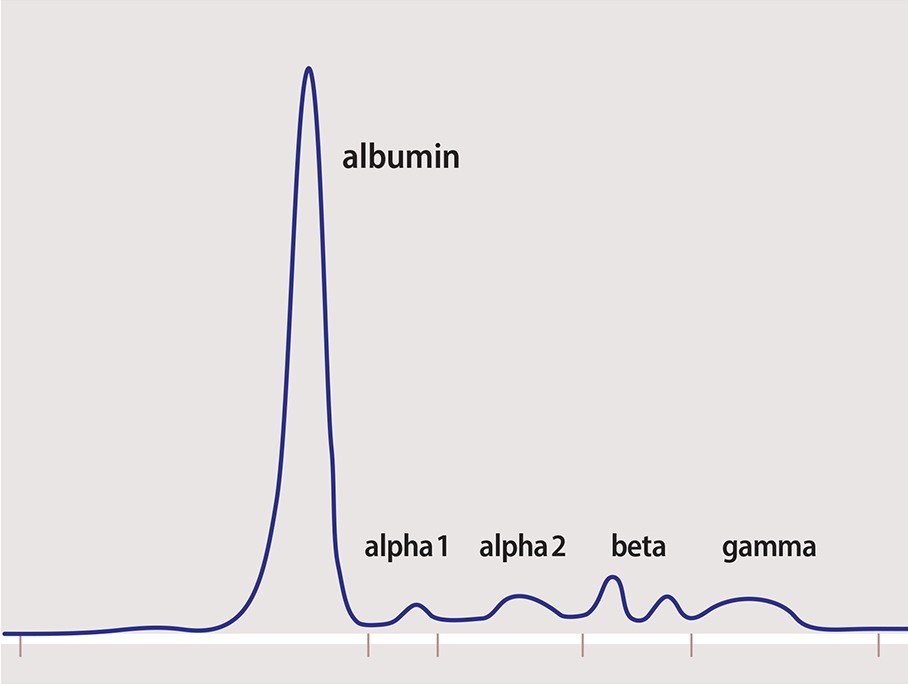
Measuring M Protein Spep Upep Tests Int L Myeloma Fdn

Protein Assay Lab Sudhapetluri
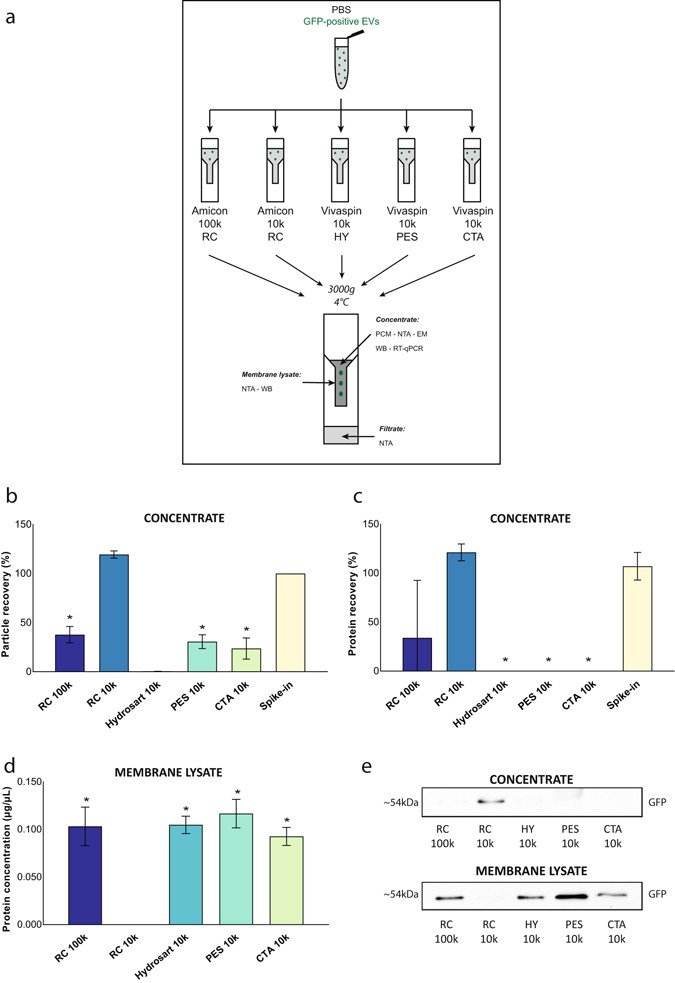
Confounding Factors Of Ultrafiltration And Protein Analysis In Extracellular Vesicle Research Scientific Reports

R For Biochemists Drawing The Protein Assay With Ggplot

In Vitro Protein Releasing Assay Graph Shows The Protein Concentration Download Scientific Diagram
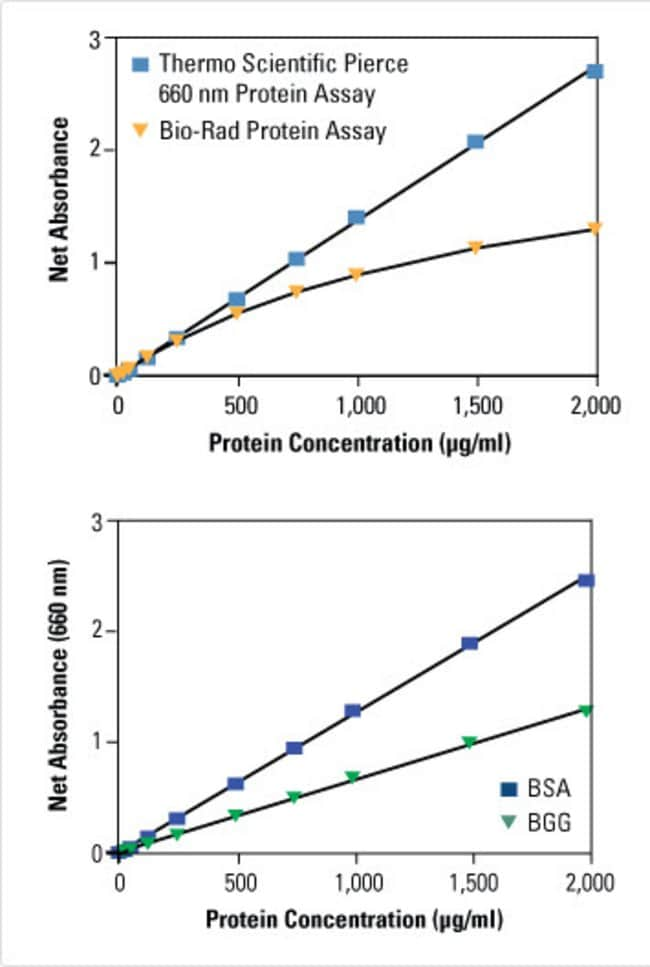
Pierce 660nm Protein Assay Reagent

Team Sdu Denmark Experiments 16 Igem Org

Figure 1 From Linearization Of The Bradford Protein Assay Semantic Scholar
Http Datasheets Scbt Com Sc 23 Pdf
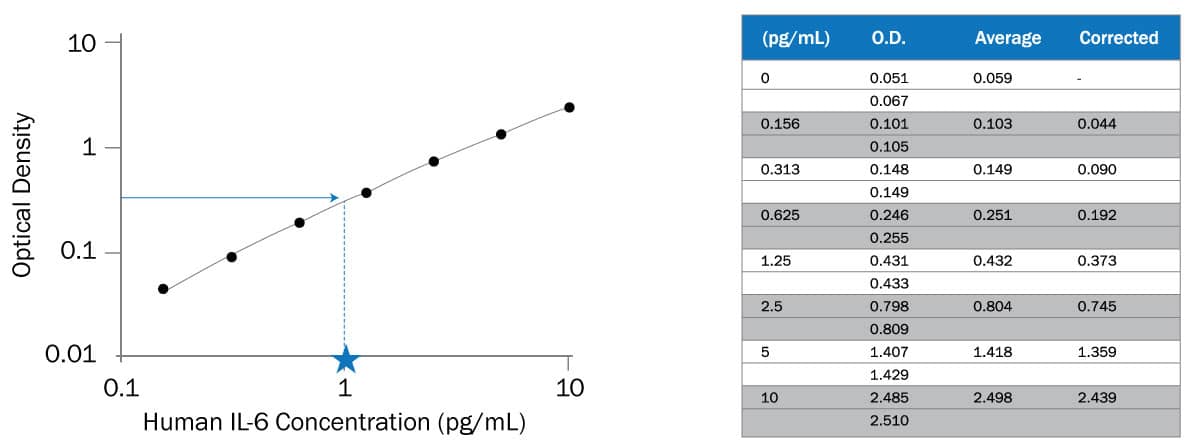
How To Analyze Elisa Data R D Systems
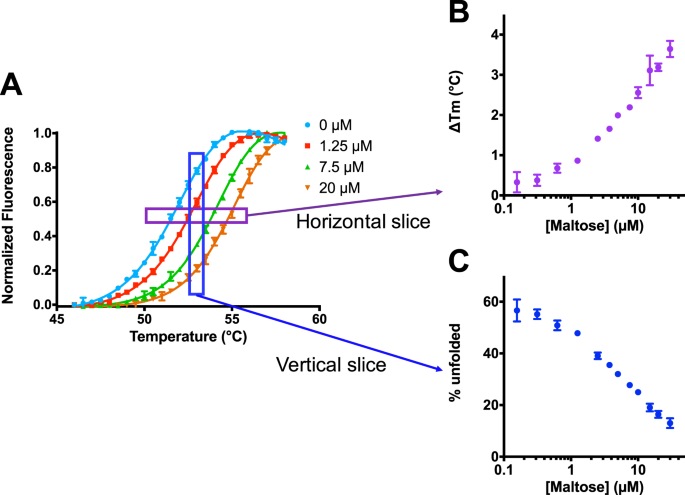
Isothermal Analysis Of Thermofluor Data Can Readily Provide Quantitative Binding Affinities Scientific Reports

Linear Range Of Protein Quantification For The Lowry And Bradford Download Scientific Diagram

User Keyun Wang Notebook Experimental Biological Chemistry I 12 11 07 Openwetware

Standard Curve An Overview Sciencedirect Topics
Http Himedialabs Com Td Htbc005 Pdf

Protein Quantitation

Bradford Protein Assay Lab Adrian Biology
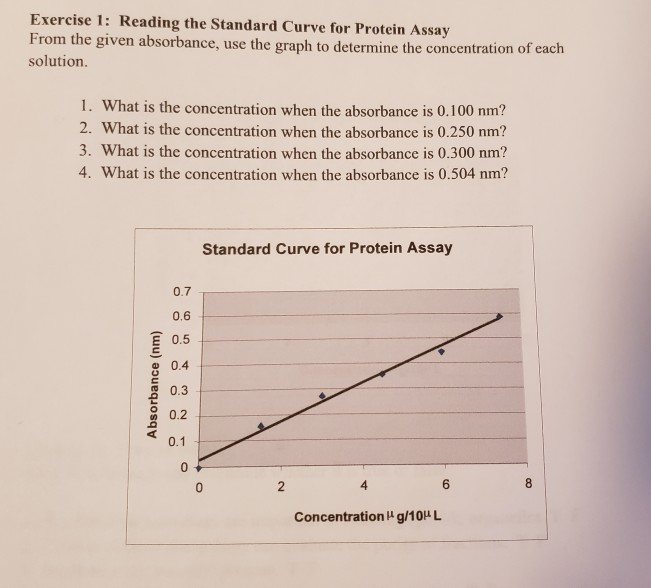
Solved Exercise 1 Reading The Standard Curve For Protein Chegg Com
Q Tbn 3aand9gcqpvst Dwjmf0cig7j8wdewswqbb2pje5ywxdeave9j8y8nug44 Usqp Cau



